Fang Wu
LatentChem: From Textual CoT to Latent Thinking in Chemical Reasoning
Feb 06, 2026Abstract:Chemical large language models (LLMs) predominantly rely on explicit Chain-of-Thought (CoT) in natural language to perform complex reasoning. However, chemical reasoning is inherently continuous and structural, and forcing it into discrete linguistic tokens introduces a fundamental representation mismatch that constrains both efficiency and performance. We introduce LatentChem, a latent reasoning interface that decouples chemical computation from textual generation, enabling models to perform multi-step reasoning directly in continuous latent space while emitting language only for final outputs. Remarkably, we observe a consistent emergent behavior: when optimized solely for task success, models spontaneously internalize reasoning, progressively abandoning verbose textual derivations in favor of implicit latent computation. This shift is not merely stylistic but computationally advantageous. Across diverse chemical reasoning benchmarks, LatentChem achieves a 59.88\% non-tie win rate over strong CoT-based baselines on ChemCoTBench, while delivering a 10.84$\times$ average inference speedup. Our results provide empirical evidence that chemical reasoning is more naturally and effectively realized as continuous latent dynamics rather than discretized linguistic trajectories.
Molecular Representations in Implicit Functional Space via Hyper-Networks
Jan 29, 2026Abstract:Molecular representations fundamentally shape how machine learning systems reason about molecular structure and physical properties. Most existing approaches adopt a discrete pipeline: molecules are encoded as sequences, graphs, or point clouds, mapped to fixed-dimensional embeddings, and then used for task-specific prediction. This paradigm treats molecules as discrete objects, despite their intrinsically continuous and field-like physical nature. We argue that molecular learning can instead be formulated as learning in function space. Specifically, we model each molecule as a continuous function over three-dimensional (3D) space and treat this molecular field as the primary object of representation. From this perspective, conventional molecular representations arise as particular sampling schemes of an underlying continuous object. We instantiate this formulation with MolField, a hyper-network-based framework that learns distributions over molecular fields. To ensure physical consistency, these functions are defined over canonicalized coordinates, yielding invariance to global SE(3) transformations. To enable learning directly over functions, we introduce a structured weight tokenization and train a sequence-based hyper-network to model a shared prior over molecular fields. We evaluate MolField on molecular dynamics and property prediction. Our results show that treating molecules as continuous functions fundamentally changes how molecular representations generalize across tasks and yields downstream behavior that is stable to how molecules are discretized or queried.
Why Reasoning Fails to Plan: A Planning-Centric Analysis of Long-Horizon Decision Making in LLM Agents
Jan 29, 2026Abstract:Large language model (LLM)-based agents exhibit strong step-by-step reasoning capabilities over short horizons, yet often fail to sustain coherent behavior over long planning horizons. We argue that this failure reflects a fundamental mismatch: step-wise reasoning induces a form of step-wise greedy policy that is adequate for short horizons but fails in long-horizon planning, where early actions must account for delayed consequences. From this planning-centric perspective, we study LLM-based agents in deterministic, fully structured environments with explicit state transitions and evaluation signals. Our analysis reveals a core failure mode of reasoning-based policies: locally optimal choices induced by step-wise scoring lead to early myopic commitments that are systematically amplified over time and difficult to recover from. We introduce FLARE (Future-aware Lookahead with Reward Estimation) as a minimal instantiation of future-aware planning to enforce explicit lookahead, value propagation, and limited commitment in a single model, allowing downstream outcomes to influence early decisions. Across multiple benchmarks, agent frameworks, and LLM backbones, FLARE consistently improves task performance and planning-level behavior, frequently allowing LLaMA-8B with FLARE to outperform GPT-4o with standard step-by-step reasoning. These results establish a clear distinction between reasoning and planning.
Surface-based Molecular Design with Multi-modal Flow Matching
Jan 08, 2026Abstract:Therapeutic peptides show promise in targeting previously undruggable binding sites, with recent advancements in deep generative models enabling full-atom peptide co-design for specific protein receptors. However, the critical role of molecular surfaces in protein-protein interactions (PPIs) has been underexplored. To bridge this gap, we propose an omni-design peptides generation paradigm, called SurfFlow, a novel surface-based generative algorithm that enables comprehensive co-design of sequence, structure, and surface for peptides. SurfFlow employs a multi-modality conditional flow matching (CFM) architecture to learn distributions of surface geometries and biochemical properties, enhancing peptide binding accuracy. Evaluated on the comprehensive PepMerge benchmark, SurfFlow consistently outperforms full-atom baselines across all metrics. These results highlight the advantages of considering molecular surfaces in de novo peptide discovery and demonstrate the potential of integrating multiple protein modalities for more effective therapeutic peptide discovery.
A Semi-supervised Molecular Learning Framework for Activity Cliff Estimation
Jan 08, 2026Abstract:Machine learning (ML) enables accurate and fast molecular property predictions, which are of interest in drug discovery and material design. Their success is based on the principle of similarity at its heart, assuming that similar molecules exhibit close properties. However, activity cliffs challenge this principle, and their presence leads to a sharp decline in the performance of existing ML algorithms, particularly graph-based methods. To overcome this obstacle under a low-data scenario, we propose a novel semi-supervised learning (SSL) method dubbed SemiMol, which employs predictions on numerous unannotated data as pseudo-signals for subsequent training. Specifically, we introduce an additional instructor model to evaluate the accuracy and trustworthiness of proxy labels because existing pseudo-labeling approaches require probabilistic outputs to reveal the model's confidence and fail to be applied in regression tasks. Moreover, we design a self-adaptive curriculum learning algorithm to progressively move the target model toward hard samples at a controllable pace. Extensive experiments on 30 activity cliff datasets demonstrate that SemiMol significantly enhances graph-based ML architectures and outpasses state-of-the-art pretraining and SSL baselines.
Toward Global Large Language Models in Medicine
Jan 05, 2026Abstract:Despite continuous advances in medical technology, the global distribution of health care resources remains uneven. The development of large language models (LLMs) has transformed the landscape of medicine and holds promise for improving health care quality and expanding access to medical information globally. However, existing LLMs are primarily trained on high-resource languages, limiting their applicability in global medical scenarios. To address this gap, we constructed GlobMed, a large multilingual medical dataset, containing over 500,000 entries spanning 12 languages, including four low-resource languages. Building on this, we established GlobMed-Bench, which systematically assesses 56 state-of-the-art proprietary and open-weight LLMs across multiple multilingual medical tasks, revealing significant performance disparities across languages, particularly for low-resource languages. Additionally, we introduced GlobMed-LLMs, a suite of multilingual medical LLMs trained on GlobMed, with parameters ranging from 1.7B to 8B. GlobMed-LLMs achieved an average performance improvement of over 40% relative to baseline models, with a more than threefold increase in performance on low-resource languages. Together, these resources provide an important foundation for advancing the equitable development and application of LLMs globally, enabling broader language communities to benefit from technological advances.
From Supervision to Exploration: What Does Protein Language Model Learn During Reinforcement Learning?
Oct 02, 2025Abstract:Protein language models (PLMs) have advanced computational protein science through large-scale pretraining and scalable architectures. In parallel, reinforcement learning (RL) has broadened exploration and enabled precise multi-objective optimization in protein design. Yet whether RL can push PLMs beyond their pretraining priors to uncover latent sequence-structure-function rules remains unclear. We address this by pairing RL with PLMs across four domains: antimicrobial peptide design, kinase variant optimization, antibody engineering, and inverse folding. Using diverse RL algorithms and model classes, we ask if RL improves sampling efficiency and, more importantly, if it reveals capabilities not captured by supervised learning. Across benchmarks, RL consistently boosts success rates and sample efficiency. Performance follows a three-factor interaction: task headroom, reward fidelity, and policy capacity jointly determine gains. When rewards are accurate and informative, policies have sufficient capacity, and tasks leave room beyond supervised baselines, improvements scale; when rewards are noisy or capacity is constrained, gains saturate despite exploration. This view yields practical guidance for RL in protein design: prioritize reward modeling and calibration before scaling policy size, match algorithm and regularization strength to task difficulty, and allocate capacity where marginal gains are largest. Implementation is available at https://github.com/chq1155/RL-PLM.
Agent KB: Leveraging Cross-Domain Experience for Agentic Problem Solving
Jul 08, 2025Abstract:As language agents tackle increasingly complex tasks, they struggle with effective error correction and experience reuse across domains. We introduce Agent KB, a hierarchical experience framework that enables complex agentic problem solving via a novel Reason-Retrieve-Refine pipeline. Agent KB addresses a core limitation: agents traditionally cannot learn from each other's experiences. By capturing both high-level strategies and detailed execution logs, Agent KB creates a shared knowledge base that enables cross-agent knowledge transfer. Evaluated on the GAIA benchmark, Agent KB improves success rates by up to 16.28 percentage points. On the most challenging tasks, Claude-3 improves from 38.46% to 57.69%, while GPT-4 improves from 53.49% to 73.26% on intermediate tasks. On SWE-bench code repair, Agent KB enables Claude-3 to improve from 41.33% to 53.33%. Our results suggest that Agent KB provides a modular, framework-agnostic infrastructure for enabling agents to learn from past experiences and generalize successful strategies to new tasks.
When to Trust Context: Self-Reflective Debates for Context Reliability
Jun 06, 2025


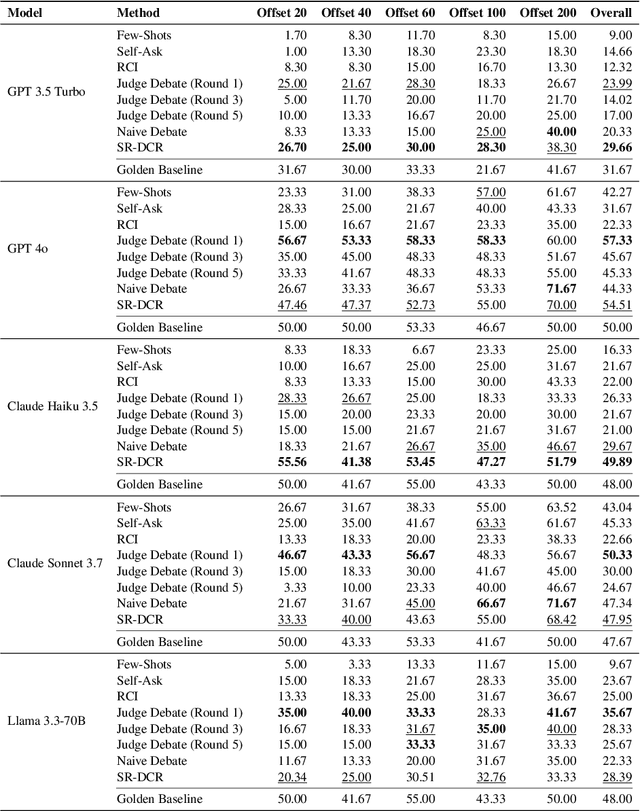
Abstract:Large language models frequently encounter conflicts between their parametric knowledge and contextual input, often resulting in factual inconsistencies or hallucinations. We propose Self-Reflective Debate for Contextual Reliability (SR-DCR), a lightweight framework that integrates token-level self-confidence with an asymmetric multi-agent debate to adjudicate such conflicts. A critic, deprived of context, challenges a defender who argues from the given passage; a judge model evaluates the debate and determines the context's reliability. The final answer is selected by combining the verdict with model confidence. Experiments on the ClashEval benchmark demonstrate that SR-DCR consistently enhances robustness to misleading context while maintaining accuracy on trustworthy inputs, outperforming both classical debate and confidence-only baselines with minimal computational overhead. The code is available at https://github.com/smiles724/Self-Reflective-Debates.
Large Language Models are Good Relational Learners
Jun 06, 2025



Abstract:Large language models (LLMs) have demonstrated remarkable capabilities across various domains, yet their application to relational deep learning (RDL) remains underexplored. Existing approaches adapt LLMs by traversing relational links between entities in a database and converting the structured data into flat text documents. Still, this text-based serialization disregards critical relational structures, introduces redundancy, and often exceeds standard LLM context lengths. We introduce Rel-LLM, a novel architecture that utilizes a graph neural network (GNN)- based encoder to generate structured relational prompts for LLMs within a retrieval-augmented generation (RAG) framework. Unlike traditional text-based serialization approaches, our method preserves the inherent relational structure of databases while enabling LLMs to effectively process and reason over complex entity relationships. Specifically, the GNN encoder extracts a local subgraph around an entity to build feature representations that contain relevant entity relationships and temporal dependencies. These representations are transformed into structured prompts using a denormalization process, effectively allowing the LLM to reason over relational structures. Through extensive experiments, we demonstrate that Rel-LLM outperforms existing methods on key RDL tasks, offering a scalable and efficient approach to integrating LLMs with structured data sources. Code is available at https://github.com/smiles724/Rel-LLM.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge