Jin Han
Seeing Through the Rain: Resolving High-Frequency Conflicts in Deraining and Super-Resolution via Diffusion Guidance
Nov 16, 2025Abstract:Clean images are crucial for visual tasks such as small object detection, especially at high resolutions. However, real-world images are often degraded by adverse weather, and weather restoration methods may sacrifice high-frequency details critical for analyzing small objects. A natural solution is to apply super-resolution (SR) after weather removal to recover both clarity and fine structures. However, simply cascading restoration and SR struggle to bridge their inherent conflict: removal aims to remove high-frequency weather-induced noise, while SR aims to hallucinate high-frequency textures from existing details, leading to inconsistent restoration contents. In this paper, we take deraining as a case study and propose DHGM, a Diffusion-based High-frequency Guided Model for generating clean and high-resolution images. DHGM integrates pre-trained diffusion priors with high-pass filters to simultaneously remove rain artifacts and enhance structural details. Extensive experiments demonstrate that DHGM achieves superior performance over existing methods, with lower costs.
PoseX: AI Defeats Physics Approaches on Protein-Ligand Cross Docking
May 03, 2025Abstract:Recently, significant progress has been made in protein-ligand docking, especially in modern deep learning methods, and some benchmarks were proposed, e.g., PoseBench, Plinder. However, these benchmarks suffer from less practical evaluation setups (e.g., blind docking, self docking), or heavy framework that involves training, raising challenges to assess docking methods efficiently. To fill this gap, we proposed PoseX, an open-source benchmark focusing on self-docking and cross-docking, to evaluate the algorithmic advances practically and comprehensively. Specifically, first, we curate a new evaluation dataset with 718 entries for self docking and 1,312 for cross docking; second, we incorporate 22 docking methods across three methodological categories, including (1) traditional physics-based methods (e.g., Schr\"odinger Glide), (2) AI docking methods (e.g., DiffDock), (3) AI co-folding methods (e.g., AlphaFold3); third, we design a relaxation method as post-processing to minimize conformation energy and refine binding pose; fourth, we released a leaderboard to rank submitted models in real time. We draw some key insights via extensive experiments: (1) AI-based approaches have already surpassed traditional physics-based approaches in overall docking accuracy (RMSD). The longstanding generalization issues that have plagued AI molecular docking have been significantly alleviated in the latest models. (2) The stereochemical deficiencies of AI-based approaches can be greatly alleviated with post-processing relaxation. Combining AI docking methods with the enhanced relaxation method achieves the best performance to date. (3) AI co-folding methods commonly face ligand chirality issues, which cannot be resolved by relaxation. The code, curated dataset and leaderboard are released at https://github.com/CataAI/PoseX.
Hashing for Protein Structure Similarity Search
Nov 13, 2024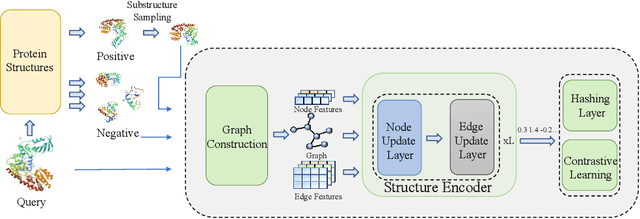
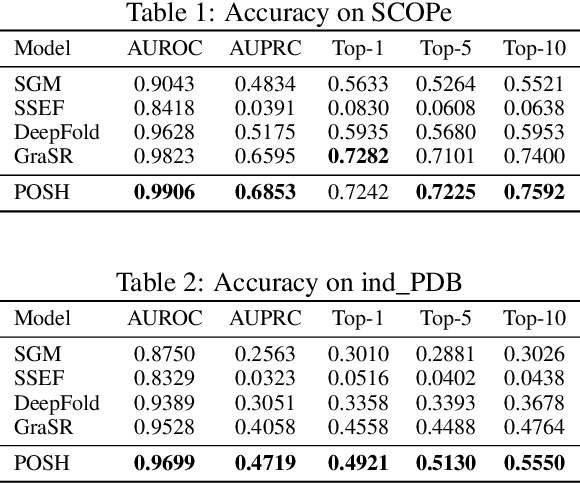
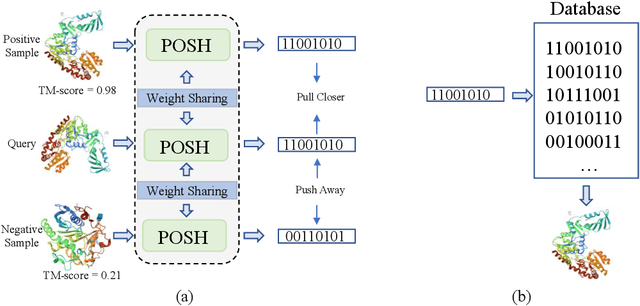
Abstract:Protein structure similarity search (PSSS), which tries to search proteins with similar structures, plays a crucial role across diverse domains from drug design to protein function prediction and molecular evolution. Traditional alignment-based PSSS methods, which directly calculate alignment on the protein structures, are highly time-consuming with high memory cost. Recently, alignment-free methods, which represent protein structures as fixed-length real-valued vectors, are proposed for PSSS. Although these methods have lower time and memory cost than alignment-based methods, their time and memory cost is still too high for large-scale PSSS, and their accuracy is unsatisfactory. In this paper, we propose a novel method, called $\underline{\text{p}}$r$\underline{\text{o}}$tein $\underline{\text{s}}$tructure $\underline{\text{h}}$ashing (POSH), for PSSS. POSH learns a binary vector representation for each protein structure, which can dramatically reduce the time and memory cost for PSSS compared with real-valued vector representation based methods. Furthermore, in POSH we also propose expressive hand-crafted features and a structure encoder to well model both node and edge interactions in proteins. Experimental results on real datasets show that POSH can outperform other methods to achieve state-of-the-art accuracy. Furthermore, POSH achieves a memory saving of more than six times and speed improvement of more than four times, compared with other methods.
Hashing based Contrastive Learning for Virtual Screening
Jul 29, 2024Abstract:Virtual screening (VS) is a critical step in computer-aided drug discovery, aiming to identify molecules that bind to a specific target receptor like protein. Traditional VS methods, such as docking, are often too time-consuming for screening large-scale molecular databases. Recent advances in deep learning have demonstrated that learning vector representations for both proteins and molecules using contrastive learning can outperform traditional docking methods. However, given that target databases often contain billions of molecules, real-valued vector representations adopted by existing methods can still incur significant memory and time costs in VS. To address this problem, in this paper we propose a hashing-based contrastive learning method, called DrugHash, for VS. DrugHash treats VS as a retrieval task that uses efficient binary hash codes for retrieval. In particular, DrugHash designs a simple yet effective hashing strategy to enable end-to-end learning of binary hash codes for both protein and molecule modalities, which can dramatically reduce the memory and time costs with higher accuracy compared with existing methods. Experimental results show that DrugHash can outperform existing methods to achieve state-of-the-art accuracy, with a memory saving of 32$\times$ and a speed improvement of 3.5$\times$.
QuakeBERT: Accurate Classification of Social Media Texts for Rapid Earthquake Impact Assessment
May 06, 2024

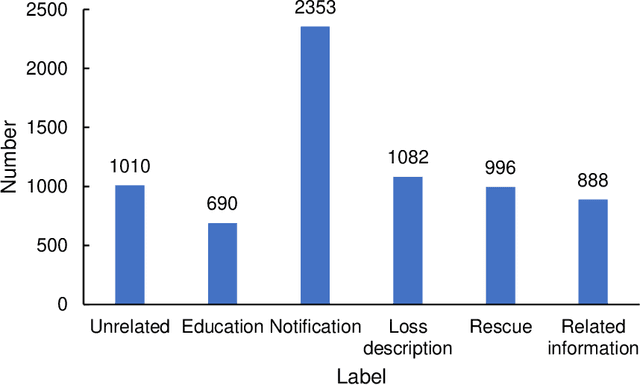
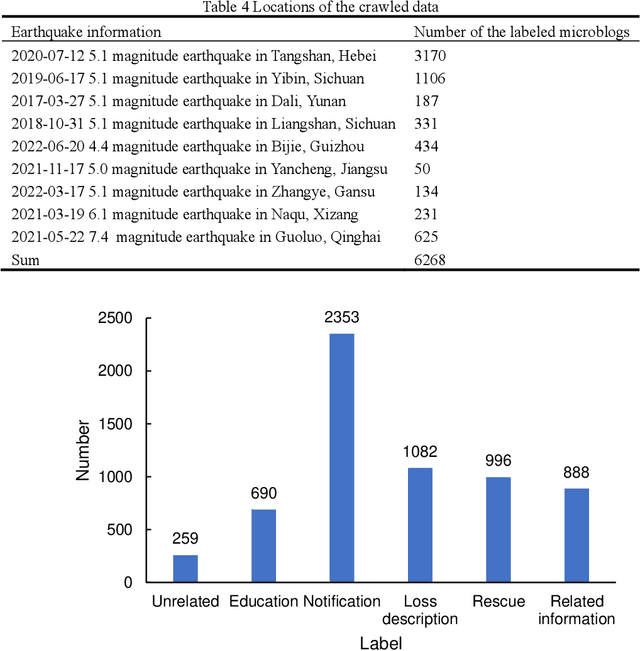
Abstract:Social media aids disaster response but suffers from noise, hindering accurate impact assessment and decision making for resilient cities, which few studies considered. To address the problem, this study proposes the first domain-specific LLM model and an integrated method for rapid earthquake impact assessment. First, a few categories are introduced to classify and filter microblogs considering their relationship to the physical and social impacts of earthquakes, and a dataset comprising 7282 earthquake-related microblogs from twenty earthquakes in different locations is developed as well. Then, with a systematic analysis of various influential factors, QuakeBERT, a domain-specific large language model (LLM), is developed and fine-tuned for accurate classification and filtering of microblogs. Meanwhile, an integrated method integrating public opinion trend analysis, sentiment analysis, and keyword-based physical impact quantification is introduced to assess both the physical and social impacts of earthquakes based on social media texts. Experiments show that data diversity and data volume dominate the performance of QuakeBERT and increase the macro average F1 score by 27%, while the best classification model QuakeBERT outperforms the CNN- or RNN-based models by improving the macro average F1 score from 60.87% to 84.33%. Finally, the proposed approach is applied to assess two earthquakes with the same magnitude and focal depth. Results show that the proposed approach can effectively enhance the impact assessment process by accurate detection of noisy microblogs, which enables effective post-disaster emergency responses to create more resilient cities.
Learning Differential Diagnosis of Skin Conditions with Co-occurrence Supervision using Graph Convolutional Networks
Jul 13, 2020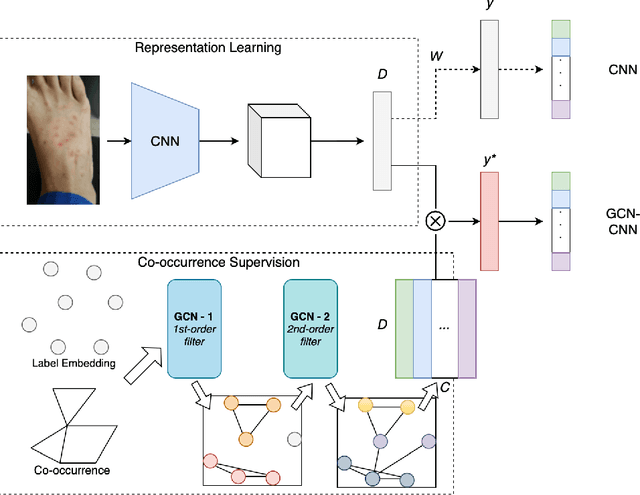
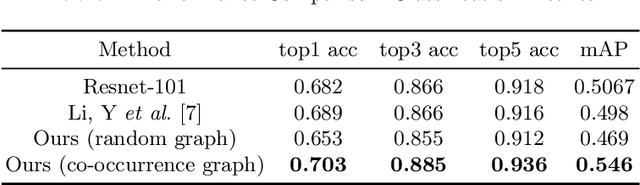


Abstract:Skin conditions are reported the 4th leading cause of nonfatal disease burden worldwide. However, given the colossal spectrum of skin disorders defined clinically and shortage in dermatology expertise, diagnosing skin conditions in a timely and accurate manner remains a challenging task. Using computer vision technologies, a deep learning system has proven effective assisting clinicians in image diagnostics of radiology, ophthalmology and more. In this paper, we propose a deep learning system (DLS) that may predict differential diagnosis of skin conditions using clinical images. Our DLS formulates the differential diagnostics as a multi-label classification task over 80 conditions when only incomplete image labels are available. We tackle the label incompleteness problem by combining a classification network with a Graph Convolutional Network (GCN) that characterizes label co-occurrence and effectively regularizes it towards a sparse representation. Our approach is demonstrated on 136,462 clinical images and concludes that the classification accuracy greatly benefit from the Co-occurrence supervision. Our DLS achieves 93.6% top-5 accuracy on 12,378 test images and consistently outperform the baseline classification network.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge