Stan Z. Li
Departures: Distributional Transport for Single-Cell Perturbation Prediction with Neural Schrödinger Bridges
Nov 17, 2025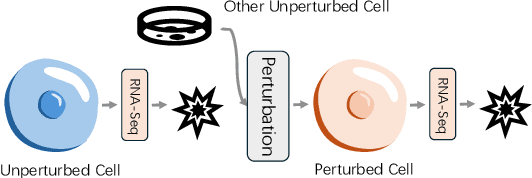
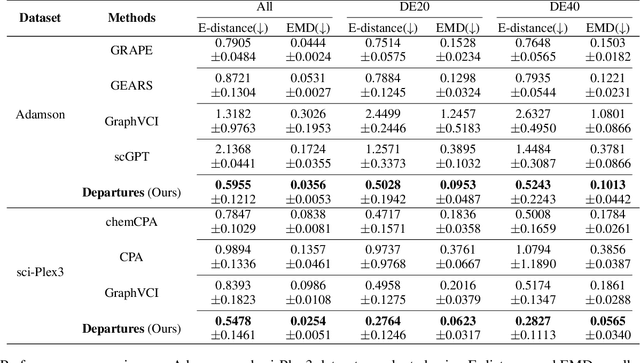
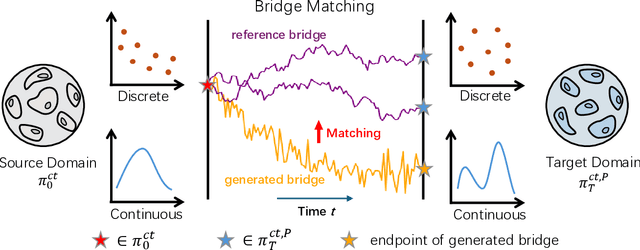

Abstract:Predicting single-cell perturbation outcomes directly advances gene function analysis and facilitates drug candidate selection, making it a key driver of both basic and translational biomedical research. However, a major bottleneck in this task is the unpaired nature of single-cell data, as the same cell cannot be observed both before and after perturbation due to the destructive nature of sequencing. Although some neural generative transport models attempt to tackle unpaired single-cell perturbation data, they either lack explicit conditioning or depend on prior spaces for indirect distribution alignment, limiting precise perturbation modeling. In this work, we approximate Schrödinger Bridge (SB), which defines stochastic dynamic mappings recovering the entropy-regularized optimal transport (OT), to directly align the distributions of control and perturbed single-cell populations across different perturbation conditions. Unlike prior SB approximations that rely on bidirectional modeling to infer optimal source-target sample coupling, we leverage Minibatch-OT based pairing to avoid such bidirectional inference and the associated ill-posedness of defining the reverse process. This pairing directly guides bridge learning, yielding a scalable approximation to the SB. We approximate two SB models, one modeling discrete gene activation states and the other continuous expression distributions. Joint training enables accurate perturbation modeling and captures single-cell heterogeneity. Experiments on public genetic and drug perturbation datasets show that our model effectively captures heterogeneous single-cell responses and achieves state-of-the-art performance.
MergeDNA: Context-aware Genome Modeling with Dynamic Tokenization through Token Merging
Nov 17, 2025



Abstract:Modeling genomic sequences faces two unsolved challenges: the information density varies widely across different regions, while there is no clearly defined minimum vocabulary unit. Relying on either four primitive bases or independently designed DNA tokenizers, existing approaches with naive masked language modeling pre-training often fail to adapt to the varying complexities of genomic sequences. Leveraging Token Merging techniques, this paper introduces a hierarchical architecture that jointly optimizes a dynamic genomic tokenizer and latent Transformers with context-aware pre-training tasks. As for network structures, the tokenization module automatically chunks adjacent bases into words by stacking multiple layers of the differentiable token merging blocks with local-window constraints, then a Latent Encoder captures the global context of these merged words by full-attention blocks. Symmetrically employing a Latent Decoder and a Local Decoder, MergeDNA learns with two pre-training tasks: Merged Token Reconstruction simultaneously trains the dynamic tokenization module and adaptively filters important tokens, while Adaptive Masked Token Modeling learns to predict these filtered tokens to capture informative contents. Extensive experiments show that MergeDNA achieves superior performance on three popular DNA benchmarks and several multi-omics tasks with fine-tuning or zero-shot evaluation, outperforming typical tokenization methods and large-scale DNA foundation models.
AlphaFold Database Debiasing for Robust Inverse Folding
Jun 10, 2025Abstract:The AlphaFold Protein Structure Database (AFDB) offers unparalleled structural coverage at near-experimental accuracy, positioning it as a valuable resource for data-driven protein design. However, its direct use in training deep models that are sensitive to fine-grained atomic geometry, such as inverse folding, exposes a critical limitation. Comparative analysis of structural feature distributions reveals that AFDB structures exhibit distinct statistical regularities, reflecting a systematic geometric bias that deviates from the conformational diversity found in experimentally determined structures from the Protein Data Bank (PDB). While AFDB structures are cleaner and more idealized, PDB structures capture the intrinsic variability and physical realism essential for generalization in downstream tasks. To address this discrepancy, we introduce a Debiasing Structure AutoEncoder (DeSAE) that learns to reconstruct native-like conformations from intentionally corrupted backbone geometries. By training the model to recover plausible structural states, DeSAE implicitly captures a more robust and natural structural manifold. At inference, applying DeSAE to AFDB structures produces debiased structures that significantly improve inverse folding performance across multiple benchmarks. This work highlights the critical impact of subtle systematic biases in predicted structures and presents a principled framework for debiasing, significantly boosting the performance of structure-based learning tasks like inverse folding.
Tokenizing Electron Cloud in Protein-Ligand Interaction Learning
May 25, 2025



Abstract:The affinity and specificity of protein-molecule binding directly impact functional outcomes, uncovering the mechanisms underlying biological regulation and signal transduction. Most deep-learning-based prediction approaches focus on structures of atoms or fragments. However, quantum chemical properties, such as electronic structures, are the key to unveiling interaction patterns but remain largely underexplored. To bridge this gap, we propose ECBind, a method for tokenizing electron cloud signals into quantized embeddings, enabling their integration into downstream tasks such as binding affinity prediction. By incorporating electron densities, ECBind helps uncover binding modes that cannot be fully represented by atom-level models. Specifically, to remove the redundancy inherent in electron cloud signals, a structure-aware transformer and hierarchical codebooks encode 3D binding sites enriched with electron structures into tokens. These tokenized codes are then used for specific tasks with labels. To extend its applicability to a wider range of scenarios, we utilize knowledge distillation to develop an electron-cloud-agnostic prediction model. Experimentally, ECBind demonstrates state-of-the-art performance across multiple tasks, achieving improvements of 6.42\% and 15.58\% in per-structure Pearson and Spearman correlation coefficients, respectively.
GRAPE: Heterogeneous Graph Representation Learning for Genetic Perturbation with Coding and Non-Coding Biotype
May 06, 2025Abstract:Predicting genetic perturbations enables the identification of potentially crucial genes prior to wet-lab experiments, significantly improving overall experimental efficiency. Since genes are the foundation of cellular life, building gene regulatory networks (GRN) is essential to understand and predict the effects of genetic perturbations. However, current methods fail to fully leverage gene-related information, and solely rely on simple evaluation metrics to construct coarse-grained GRN. More importantly, they ignore functional differences between biotypes, limiting the ability to capture potential gene interactions. In this work, we leverage pre-trained large language model and DNA sequence model to extract features from gene descriptions and DNA sequence data, respectively, which serve as the initialization for gene representations. Additionally, we introduce gene biotype information for the first time in genetic perturbation, simulating the distinct roles of genes with different biotypes in regulating cellular processes, while capturing implicit gene relationships through graph structure learning (GSL). We propose GRAPE, a heterogeneous graph neural network (HGNN) that leverages gene representations initialized with features from descriptions and sequences, models the distinct roles of genes with different biotypes, and dynamically refines the GRN through GSL. The results on publicly available datasets show that our method achieves state-of-the-art performance.
Adversarial Curriculum Graph-Free Knowledge Distillation for Graph Neural Networks
Apr 02, 2025Abstract:Data-free Knowledge Distillation (DFKD) is a method that constructs pseudo-samples using a generator without real data, and transfers knowledge from a teacher model to a student by enforcing the student to overcome dimensional differences and learn to mimic the teacher's outputs on these pseudo-samples. In recent years, various studies in the vision domain have made notable advancements in this area. However, the varying topological structures and non-grid nature of graph data render the methods from the vision domain ineffective. Building upon prior research into differentiable methods for graph neural networks, we propose a fast and high-quality data-free knowledge distillation approach in this paper. Without compromising distillation quality, the proposed graph-free KD method (ACGKD) significantly reduces the spatial complexity of pseudo-graphs by leveraging the Binary Concrete distribution to model the graph structure and introducing a spatial complexity tuning parameter. This approach enables efficient gradient computation for the graph structure, thereby accelerating the overall distillation process. Additionally, ACGKD eliminates the dimensional ambiguity between the student and teacher models by increasing the student's dimensions and reusing the teacher's classifier. Moreover, it equips graph knowledge distillation with a CL-based strategy to ensure the student learns graph structures progressively. Extensive experiments demonstrate that ACGKD achieves state-of-the-art performance in distilling knowledge from GNNs without training data.
Life-Code: Central Dogma Modeling with Multi-Omics Sequence Unification
Feb 11, 2025



Abstract:The interactions between DNA, RNA, and proteins are fundamental to biological processes, as illustrated by the central dogma of molecular biology. While modern biological pre-trained models have achieved great success in analyzing these macromolecules individually, their interconnected nature remains under-explored. In this paper, we follow the guidance of the central dogma to redesign both the data and model pipeline and offer a comprehensive framework, Life-Code, that spans different biological functions. As for data flow, we propose a unified pipeline to integrate multi-omics data by reverse-transcribing RNA and reverse-translating amino acids into nucleotide-based sequences. As for the model, we design a codon tokenizer and a hybrid long-sequence architecture to encode the interactions of both coding and non-coding regions with masked modeling pre-training. To model the translation and folding process with coding sequences, Life-Code learns protein structures of the corresponding amino acids by knowledge distillation from off-the-shelf protein language models. Such designs enable Life-Code to capture complex interactions within genetic sequences, providing a more comprehensive understanding of multi-omics with the central dogma. Extensive Experiments show that Life-Code achieves state-of-the-art performance on various tasks across three omics, highlighting its potential for advancing multi-omics analysis and interpretation.
A Simple yet Effective DDG Predictor is An Unsupervised Antibody Optimizer and Explainer
Feb 10, 2025Abstract:The proteins that exist today have been optimized over billions of years of natural evolution, during which nature creates random mutations and selects them. The discovery of functionally promising mutations is challenged by the limited evolutionary accessible regions, i.e., only a small region on the fitness landscape is beneficial. There have been numerous priors used to constrain protein evolution to regions of landscapes with high-fitness variants, among which the change in binding free energy (DDG) of protein complexes upon mutations is one of the most commonly used priors. However, the huge mutation space poses two challenges: (1) how to improve the efficiency of DDG prediction for fast mutation screening; and (2) how to explain mutation preferences and efficiently explore accessible evolutionary regions. To address these challenges, we propose a lightweight DDG predictor (Light-DDG), which adopts a structure-aware Transformer as the backbone and enhances it by knowledge distilled from existing powerful but computationally heavy DDG predictors. Additionally, we augmented, annotated, and released a large-scale dataset containing millions of mutation data for pre-training Light-DDG. We find that such a simple yet effective Light-DDG can serve as a good unsupervised antibody optimizer and explainer. For the target antibody, we propose a novel Mutation Explainer to learn mutation preferences, which accounts for the marginal benefit of each mutation per residue. To further explore accessible evolutionary regions, we conduct preference-guided antibody optimization and evaluate antibody candidates quickly using Light-DDG to identify desirable mutations.
G2PDiffusion: Genotype-to-Phenotype Prediction with Diffusion Models
Feb 07, 2025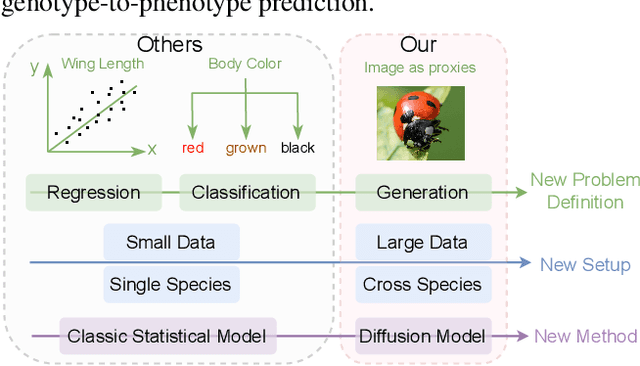
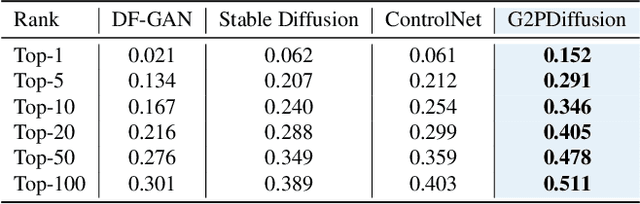
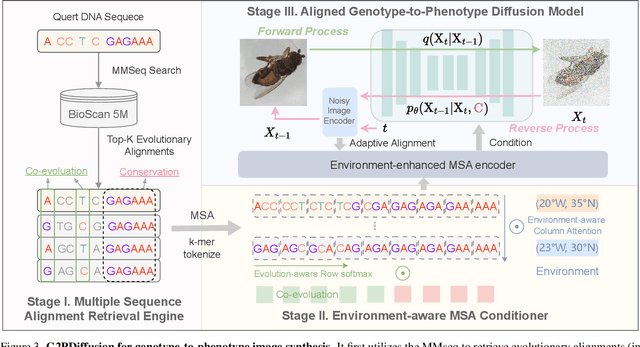
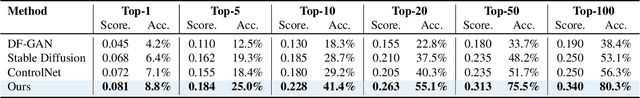
Abstract:Discovering the genotype-phenotype relationship is crucial for genetic engineering, which will facilitate advances in fields such as crop breeding, conservation biology, and personalized medicine. Current research usually focuses on single species and small datasets due to limitations in phenotypic data collection, especially for traits that require visual assessments or physical measurements. Deciphering complex and composite phenotypes, such as morphology, from genetic data at scale remains an open question. To break through traditional generic models that rely on simplified assumptions, this paper introduces G2PDiffusion, the first-of-its-kind diffusion model designed for genotype-to-phenotype generation across multiple species. Specifically, we use images to represent morphological phenotypes across species and redefine phenotype prediction as conditional image generation. To this end, this paper introduces an environment-enhanced DNA sequence conditioner and trains a stable diffusion model with a novel alignment method to improve genotype-to-phenotype consistency. Extensive experiments demonstrate that our approach enhances phenotype prediction accuracy across species, capturing subtle genetic variations that contribute to observable traits.
PhyloGen: Language Model-Enhanced Phylogenetic Inference via Graph Structure Generation
Dec 25, 2024



Abstract:Phylogenetic trees elucidate evolutionary relationships among species, but phylogenetic inference remains challenging due to the complexity of combining continuous (branch lengths) and discrete parameters (tree topology). Traditional Markov Chain Monte Carlo methods face slow convergence and computational burdens. Existing Variational Inference methods, which require pre-generated topologies and typically treat tree structures and branch lengths independently, may overlook critical sequence features, limiting their accuracy and flexibility. We propose PhyloGen, a novel method leveraging a pre-trained genomic language model to generate and optimize phylogenetic trees without dependence on evolutionary models or aligned sequence constraints. PhyloGen views phylogenetic inference as a conditionally constrained tree structure generation problem, jointly optimizing tree topology and branch lengths through three core modules: (i) Feature Extraction, (ii) PhyloTree Construction, and (iii) PhyloTree Structure Modeling. Meanwhile, we introduce a Scoring Function to guide the model towards a more stable gradient descent. We demonstrate the effectiveness and robustness of PhyloGen on eight real-world benchmark datasets. Visualization results confirm PhyloGen provides deeper insights into phylogenetic relationships.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge