Sizhe Liu
DrugAgent: Automating AI-aided Drug Discovery Programming through LLM Multi-Agent Collaboration
Nov 24, 2024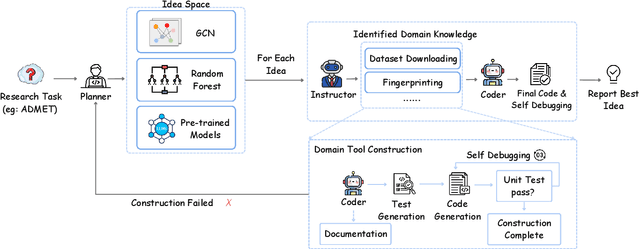
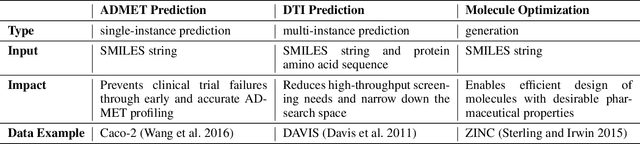
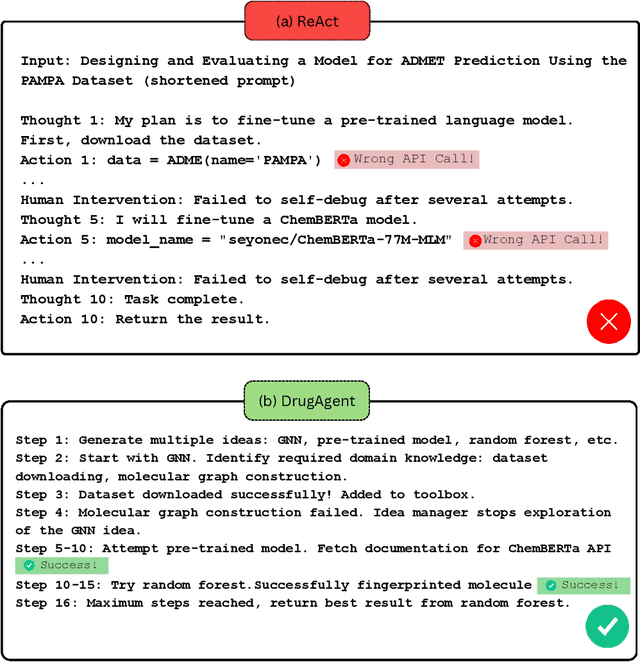
Abstract:Recent advancements in Large Language Models (LLMs) have opened new avenues for accelerating drug discovery processes. Despite their potential, several critical challenges remain unsolved, particularly in translating theoretical ideas into practical applications within the highly specialized field of pharmaceutical research, limiting practitioners from leveraging the latest AI development in drug discovery. To this end, we introduce DrugAgent, a multi-agent framework aimed at automating machine learning (ML) programming in drug discovery. DrugAgent incorporates domain expertise by identifying specific requirements and building domain-specific tools, while systematically exploring different ideas to find effective solutions. A preliminary case study demonstrates DrugAgent's potential to overcome key limitations LLMs face in drug discovery, moving toward AI-driven innovation. For example, DrugAgent is able to complete the ML programming pipeline end-to-end, from data acquisition to performance evaluation for the ADMET prediction task, and finally select the best model, where the random forest model achieves an F1 score of 0.92 when predicting absorption using the PAMPA dataset.
FlexMol: A Flexible Toolkit for Benchmarking Molecular Relational Learning
Oct 19, 2024



Abstract:Molecular relational learning (MRL) is crucial for understanding the interaction behaviors between molecular pairs, a critical aspect of drug discovery and development. However, the large feasible model space of MRL poses significant challenges to benchmarking, and existing MRL frameworks face limitations in flexibility and scope. To address these challenges, avoid repetitive coding efforts, and ensure fair comparison of models, we introduce FlexMol, a comprehensive toolkit designed to facilitate the construction and evaluation of diverse model architectures across various datasets and performance metrics. FlexMol offers a robust suite of preset model components, including 16 drug encoders, 13 protein sequence encoders, 9 protein structure encoders, and 7 interaction layers. With its easy-to-use API and flexibility, FlexMol supports the dynamic construction of over 70, 000 distinct combinations of model architectures. Additionally, we provide detailed benchmark results and code examples to demonstrate FlexMol's effectiveness in simplifying and standardizing MRL model development and comparison.
Multilingual Contrastive Decoding via Language-Agnostic Layers Skipping
Jul 15, 2024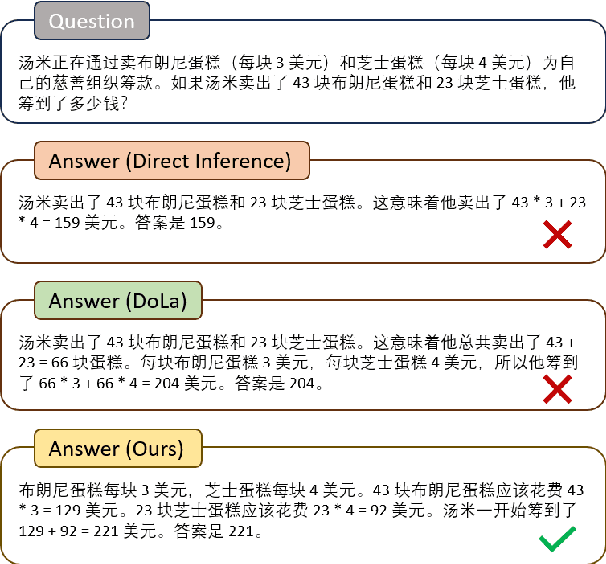
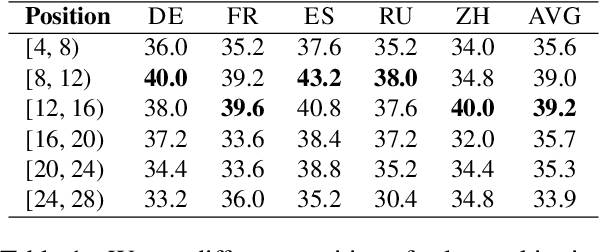
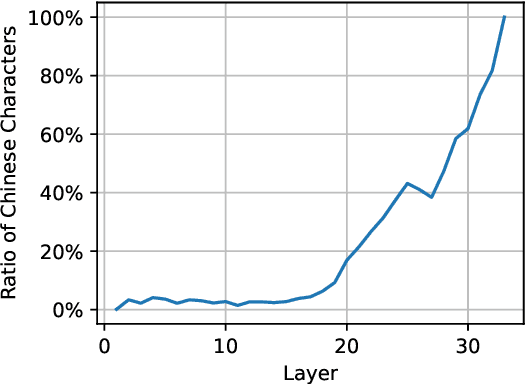
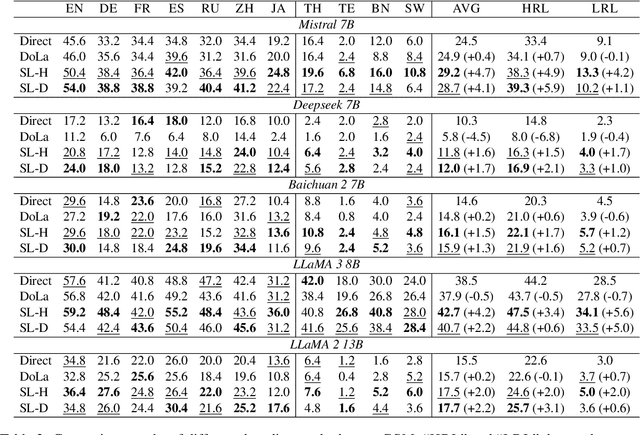
Abstract:Decoding by contrasting layers (DoLa), is designed to improve the generation quality of large language models (LLMs) by contrasting the prediction probabilities between an early exit output (amateur logits) and the final output (expert logits). However, we find that this approach does not work well on non-English tasks. Inspired by previous interpretability work on language transition during the model's forward pass, we discover that this issue arises from a language mismatch between early exit output and final output. In this work, we propose an improved contrastive decoding algorithm that is effective for diverse languages beyond English. To obtain more helpful amateur logits, we devise two strategies to skip a set of bottom, language-agnostic layers based on our preliminary analysis. Experimental results on multilingual reasoning benchmarks demonstrate that our proposed method outperforms previous contrastive decoding baselines and substantially improves LLM's chain-of-thought reasoning accuracy across 11 languages. The project will be available at: https://github.com/NJUNLP/SkipLayerCD.
Large Language Models Are Cross-Lingual Knowledge-Free Reasoners
Jun 24, 2024



Abstract:Large Language Models have demonstrated impressive reasoning capabilities across multiple languages. However, the relationship between capabilities in different languages is less explored. In this work, we decompose the process of reasoning tasks into two separated parts: knowledge retrieval and knowledge-free reasoning, and analyze the cross-lingual transferability of them. With adapted and constructed knowledge-free reasoning datasets, we show that the knowledge-free reasoning capability can be nearly perfectly transferred across various source-target language directions despite the secondary impact of resource in some specific target languages, while cross-lingual knowledge retrieval significantly hinders the transfer. Moreover, by analyzing the hidden states and feed-forward network neuron activation during the reasoning tasks, we show that higher similarity of hidden representations and larger overlap of activated neurons could explain the better cross-lingual transferability of knowledge-free reasoning than knowledge retrieval. Thus, we hypothesize that knowledge-free reasoning embeds in some language-shared mechanism, while knowledge is stored separately in different languages.
NovoBench: Benchmarking Deep Learning-based De Novo Peptide Sequencing Methods in Proteomics
Jun 16, 2024Abstract:Tandem mass spectrometry has played a pivotal role in advancing proteomics, enabling the high-throughput analysis of protein composition in biological tissues. Many deep learning methods have been developed for \emph{de novo} peptide sequencing task, i.e., predicting the peptide sequence for the observed mass spectrum. However, two key challenges seriously hinder the further advancement of this important task. Firstly, since there is no consensus for the evaluation datasets, the empirical results in different research papers are often not comparable, leading to unfair comparison. Secondly, the current methods are usually limited to amino acid-level or peptide-level precision and recall metrics. In this work, we present the first unified benchmark NovoBench for \emph{de novo} peptide sequencing, which comprises diverse mass spectrum data, integrated models, and comprehensive evaluation metrics. Recent impressive methods, including DeepNovo, PointNovo, Casanovo, InstaNovo, AdaNovo and $\pi$-HelixNovo are integrated into our framework. In addition to amino acid-level and peptide-level precision and recall, we evaluate the models' performance in terms of identifying post-tranlational modifications (PTMs), efficiency and robustness to peptide length, noise peaks and missing fragment ratio, which are important influencing factors while seldom be considered. Leveraging this benchmark, we conduct a large-scale study of current methods, report many insightful findings that open up new possibilities for future development. The benchmark will be open-sourced to facilitate future research and application.
AdaNovo: Adaptive \emph{De Novo} Peptide Sequencing with Conditional Mutual Information
Mar 15, 2024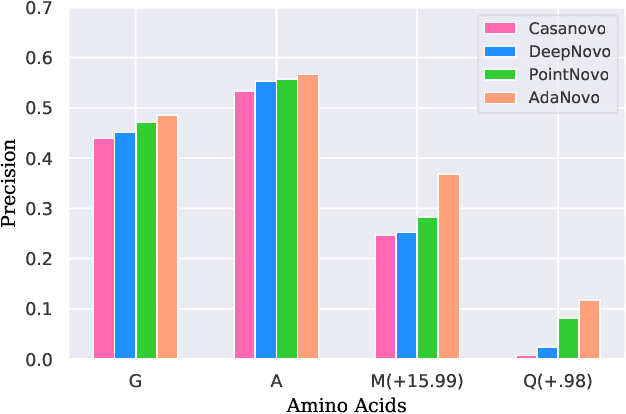
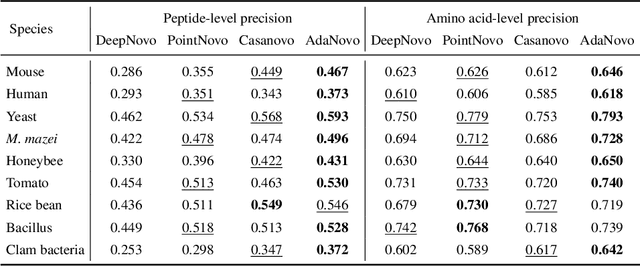
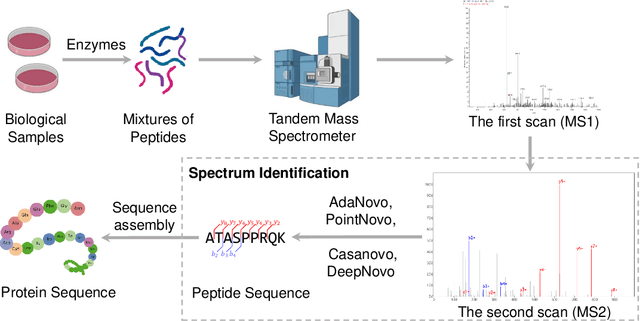
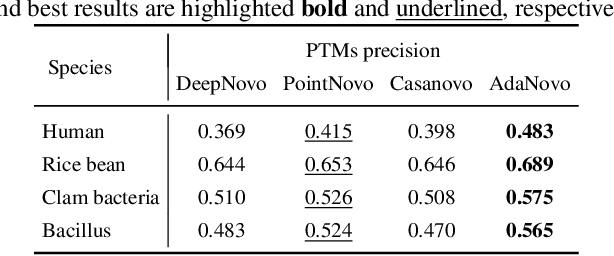
Abstract:Tandem mass spectrometry has played a pivotal role in advancing proteomics, enabling the analysis of protein composition in biological samples. Despite the development of various deep learning methods for identifying amino acid sequences (peptides) responsible for observed spectra, challenges persist in \emph{de novo} peptide sequencing. Firstly, prior methods struggle to identify amino acids with post-translational modifications (PTMs) due to their lower frequency in training data compared to canonical amino acids, further resulting in decreased peptide-level identification precision. Secondly, diverse types of noise and missing peaks in mass spectra reduce the reliability of training data (peptide-spectrum matches, PSMs). To address these challenges, we propose AdaNovo, a novel framework that calculates conditional mutual information (CMI) between the spectrum and each amino acid/peptide, using CMI for adaptive model training. Extensive experiments demonstrate AdaNovo's state-of-the-art performance on a 9-species benchmark, where the peptides in the training set are almost completely disjoint from the peptides of the test sets. Moreover, AdaNovo excels in identifying amino acids with PTMs and exhibits robustness against data noise. The supplementary materials contain the official code.
kNN-BOX: A Unified Framework for Nearest Neighbor Generation
Feb 27, 2023Abstract:Augmenting the base neural model with a token-level symbolic datastore is a novel generation paradigm and has achieved promising results in machine translation (MT). In this paper, we introduce a unified framework kNN-BOX, which enables quick development and interactive analysis for this novel paradigm. kNN-BOX decomposes the datastore-augmentation approach into three modules: datastore, retriever and combiner, thus putting diverse kNN generation methods into a unified way. Currently, kNN-BOX has provided implementation of seven popular kNN-MT variants, covering research from performance enhancement to efficiency optimization. It is easy for users to reproduce these existing works or customize their own models. Besides, users can interact with their kNN generation systems with kNN-BOX to better understand the underlying inference process in a visualized way. In the experiment section, we apply kNN-BOX for machine translation and three other seq2seq generation tasks, namely, text simplification, paraphrase generation and question generation. Experiment results show that augmenting the base neural model with kNN-BOX leads to a large performance improvement in all these tasks. The code and document of kNN-BOX is available at https://github.com/NJUNLP/knn-box.
Insights from an Industrial Collaborative Assembly Project: Lessons in Research and Collaboration
May 28, 2022



Abstract:Significant progress in robotics reveals new opportunities to advance manufacturing. Next-generation industrial automation will require both integration of distinct robotic technologies and their application to challenging industrial environments. This paper presents lessons from a collaborative assembly project between three academic research groups and an industry partner. The goal of the project is to develop a flexible, safe, and productive manufacturing cell for sub-centimeter precision assembly. Solving this problem in a high-mix, low-volume production line motivates multiple research thrusts in robotics. This work identifies new directions in collaborative robotics for industrial applications and offers insight toward strengthening collaborations between institutions in academia and industry on the development of new technologies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge