A Review of Artificial Intelligence based Biological-Tree Construction: Priorities, Methods, Applications and Trends
Paper and Code
Oct 07, 2024
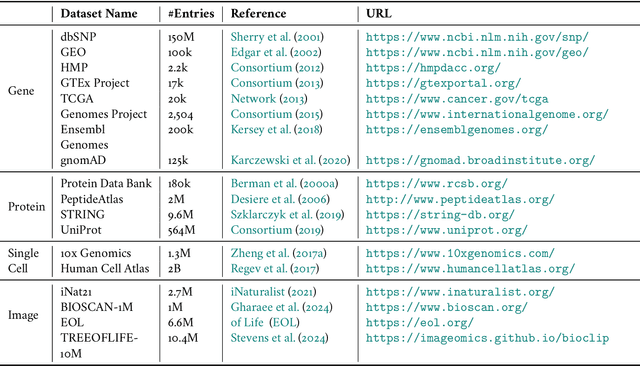
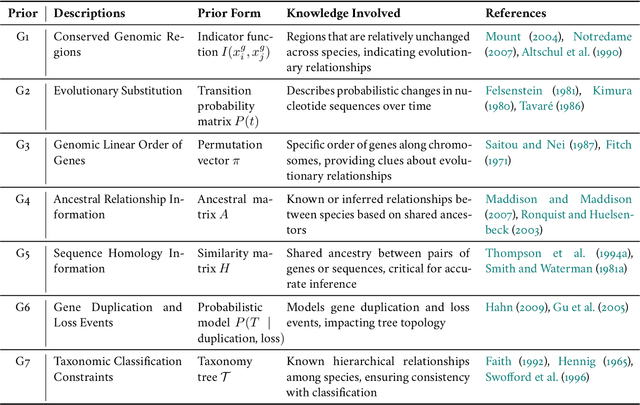

Biological tree analysis serves as a pivotal tool in uncovering the evolutionary and differentiation relationships among organisms, genes, and cells. Its applications span diverse fields including phylogenetics, developmental biology, ecology, and medicine. Traditional tree inference methods, while foundational in early studies, face increasing limitations in processing the large-scale, complex datasets generated by modern high-throughput technologies. Recent advances in deep learning offer promising solutions, providing enhanced data processing and pattern recognition capabilities. However, challenges remain, particularly in accurately representing the inherently discrete and non-Euclidean nature of biological trees. In this review, we first outline the key biological priors fundamental to phylogenetic and differentiation tree analyses, facilitating a deeper interdisciplinary understanding between deep learning researchers and biologists. We then systematically examine the commonly used data formats and databases, serving as a comprehensive resource for model testing and development. We provide a critical analysis of traditional tree generation methods, exploring their underlying biological assumptions, technical characteristics, and limitations. Current developments in deep learning-based tree generation are reviewed, highlighting both recent advancements and existing challenges. Furthermore, we discuss the diverse applications of biological trees across various biological domains. Finally, we propose potential future directions and trends in leveraging deep learning for biological tree research, aiming to guide further exploration and innovation in this field.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge