Zelin Zang
UniSurg: A Video-Native Foundation Model for Universal Understanding of Surgical Videos
Feb 05, 2026Abstract:While foundation models have advanced surgical video analysis, current approaches rely predominantly on pixel-level reconstruction objectives that waste model capacity on low-level visual details - such as smoke, specular reflections, and fluid motion - rather than semantic structures essential for surgical understanding. We present UniSurg, a video-native foundation model that shifts the learning paradigm from pixel-level reconstruction to latent motion prediction. Built on the Video Joint Embedding Predictive Architecture (V-JEPA), UniSurg introduces three key technical innovations tailored to surgical videos: 1) motion-guided latent prediction to prioritize semantically meaningful regions, 2) spatiotemporal affinity self-distillation to enforce relational consistency, and 3) feature diversity regularization to prevent representation collapse in texture-sparse surgical scenes. To enable large-scale pretraining, we curate UniSurg-15M, the largest surgical video dataset to date, comprising 3,658 hours of video from 50 sources across 13 anatomical regions. Extensive experiments across 17 benchmarks demonstrate that UniSurg significantly outperforms state-of-the-art methods on surgical workflow recognition (+14.6% F1 on EgoSurgery, +10.3% on PitVis), action triplet recognition (39.54% mAP-IVT on CholecT50), skill assessment, polyp segmentation, and depth estimation. These results establish UniSurg as a new standard for universal, motion-oriented surgical video understanding.
A Medical Multimodal Diagnostic Framework Integrating Vision-Language Models and Logic Tree Reasoning
Dec 25, 2025Abstract:With the rapid growth of large language models (LLMs) and vision-language models (VLMs) in medicine, simply integrating clinical text and medical imaging does not guarantee reliable reasoning. Existing multimodal models often produce hallucinations or inconsistent chains of thought, limiting clinical trust. We propose a diagnostic framework built upon LLaVA that combines vision-language alignment with logic-regularized reasoning. The system includes an input encoder for text and images, a projection module for cross-modal alignment, a reasoning controller that decomposes diagnostic tasks into steps, and a logic tree generator that assembles stepwise premises into verifiable conclusions. Evaluations on MedXpertQA and other benchmarks show that our method improves diagnostic accuracy and yields more interpretable reasoning traces on multimodal tasks, while remaining competitive on text-only settings. These results suggest a promising step toward trustworthy multimodal medical AI.
Anatomy-R1: Enhancing Anatomy Reasoning in Multimodal Large Language Models via Anatomical Similarity Curriculum and Group Diversity Augmentation
Dec 24, 2025Abstract:Multimodal Large Language Models (MLLMs) have achieved impressive progress in natural image reasoning, yet their potential in medical imaging remains underexplored, especially in clinical anatomical surgical images. Anatomy understanding tasks demand precise understanding and clinically coherent answers, which are difficult to achieve due to the complexity of medical data and the scarcity of high-quality expert annotations. These challenges limit the effectiveness of conventional Supervised Fine-Tuning (SFT) strategies. While recent work has demonstrated that Group Relative Policy Optimization (GRPO) can enhance reasoning in MLLMs without relying on large amounts of data, we find two weaknesses that hinder GRPO's reasoning performance in anatomy recognition: 1) knowledge cannot be effectively shared between different anatomical structures, resulting in uneven information gain and preventing the model from converging, and 2) the model quickly converges to a single reasoning path, suppressing the exploration of diverse strategies. To overcome these challenges, we propose two novel methods. First, we implement a progressive learning strategy called Anatomical Similarity Curriculum Learning by controlling question difficulty via the similarity of answer choices, enabling the model to master complex problems incrementally. Second, we utilize question augmentation referred to as Group Diversity Question Augmentation to expand the model's search space for difficult queries, mitigating the tendency to produce uniform responses. Comprehensive experiments on the SGG-VQA and OmniMedVQA benchmarks show our method achieves a significant improvement across the two benchmarks, demonstrating its effectiveness in enhancing the medical reasoning capabilities of MLLMs. The code can be found in https://github.com/tomato996/Anatomy-R1
Departures: Distributional Transport for Single-Cell Perturbation Prediction with Neural Schrödinger Bridges
Nov 17, 2025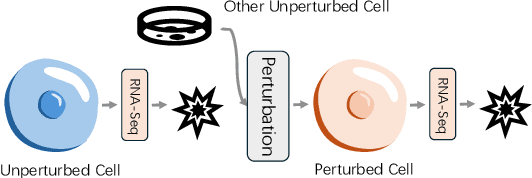
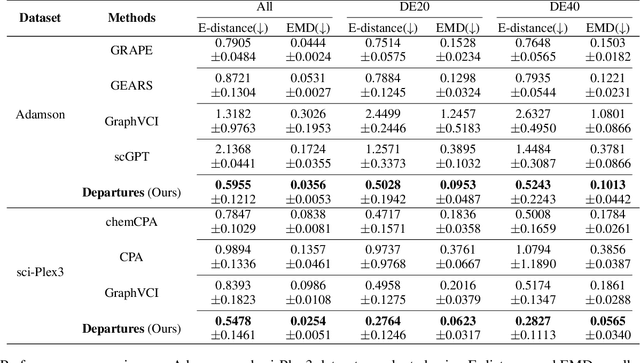
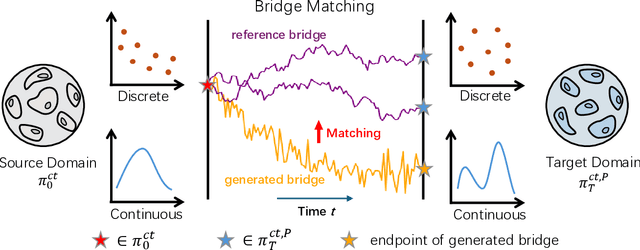

Abstract:Predicting single-cell perturbation outcomes directly advances gene function analysis and facilitates drug candidate selection, making it a key driver of both basic and translational biomedical research. However, a major bottleneck in this task is the unpaired nature of single-cell data, as the same cell cannot be observed both before and after perturbation due to the destructive nature of sequencing. Although some neural generative transport models attempt to tackle unpaired single-cell perturbation data, they either lack explicit conditioning or depend on prior spaces for indirect distribution alignment, limiting precise perturbation modeling. In this work, we approximate Schrödinger Bridge (SB), which defines stochastic dynamic mappings recovering the entropy-regularized optimal transport (OT), to directly align the distributions of control and perturbed single-cell populations across different perturbation conditions. Unlike prior SB approximations that rely on bidirectional modeling to infer optimal source-target sample coupling, we leverage Minibatch-OT based pairing to avoid such bidirectional inference and the associated ill-posedness of defining the reverse process. This pairing directly guides bridge learning, yielding a scalable approximation to the SB. We approximate two SB models, one modeling discrete gene activation states and the other continuous expression distributions. Joint training enables accurate perturbation modeling and captures single-cell heterogeneity. Experiments on public genetic and drug perturbation datasets show that our model effectively captures heterogeneous single-cell responses and achieves state-of-the-art performance.
PhyloGen: Language Model-Enhanced Phylogenetic Inference via Graph Structure Generation
Dec 25, 2024



Abstract:Phylogenetic trees elucidate evolutionary relationships among species, but phylogenetic inference remains challenging due to the complexity of combining continuous (branch lengths) and discrete parameters (tree topology). Traditional Markov Chain Monte Carlo methods face slow convergence and computational burdens. Existing Variational Inference methods, which require pre-generated topologies and typically treat tree structures and branch lengths independently, may overlook critical sequence features, limiting their accuracy and flexibility. We propose PhyloGen, a novel method leveraging a pre-trained genomic language model to generate and optimize phylogenetic trees without dependence on evolutionary models or aligned sequence constraints. PhyloGen views phylogenetic inference as a conditionally constrained tree structure generation problem, jointly optimizing tree topology and branch lengths through three core modules: (i) Feature Extraction, (ii) PhyloTree Construction, and (iii) PhyloTree Structure Modeling. Meanwhile, we introduce a Scoring Function to guide the model towards a more stable gradient descent. We demonstrate the effectiveness and robustness of PhyloGen on eight real-world benchmark datasets. Visualization results confirm PhyloGen provides deeper insights into phylogenetic relationships.
DMT-HI: MOE-based Hyperbolic Interpretable Deep Manifold Transformation for Unspervised Dimensionality Reduction
Oct 25, 2024



Abstract:Dimensionality reduction (DR) plays a crucial role in various fields, including data engineering and visualization, by simplifying complex datasets while retaining essential information. However, the challenge of balancing DR accuracy and interpretability remains crucial, particularly for users dealing with high-dimensional data. Traditional DR methods often face a trade-off between precision and transparency, where optimizing for performance can lead to reduced interpretability, and vice versa. This limitation is especially prominent in real-world applications such as image, tabular, and text data analysis, where both accuracy and interpretability are critical. To address these challenges, this work introduces the MOE-based Hyperbolic Interpretable Deep Manifold Transformation (DMT-HI). The proposed approach combines hyperbolic embeddings, which effectively capture complex hierarchical structures, with Mixture of Experts (MOE) models, which dynamically allocate tasks based on input features. DMT-HI enhances DR accuracy by leveraging hyperbolic embeddings to represent the hierarchical nature of data, while also improving interpretability by explicitly linking input data, embedding outcomes, and key features through the MOE structure. Extensive experiments demonstrate that DMT-HI consistently achieves superior performance in both DR accuracy and model interpretability, making it a robust solution for complex data analysis. The code is available at \url{https://github.com/zangzelin/code_dmthi}.
A Review of Artificial Intelligence based Biological-Tree Construction: Priorities, Methods, Applications and Trends
Oct 07, 2024
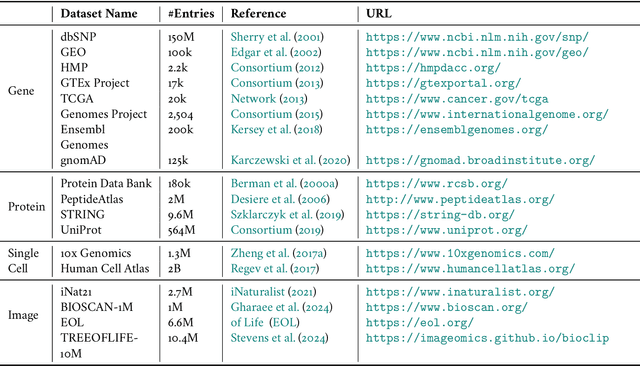
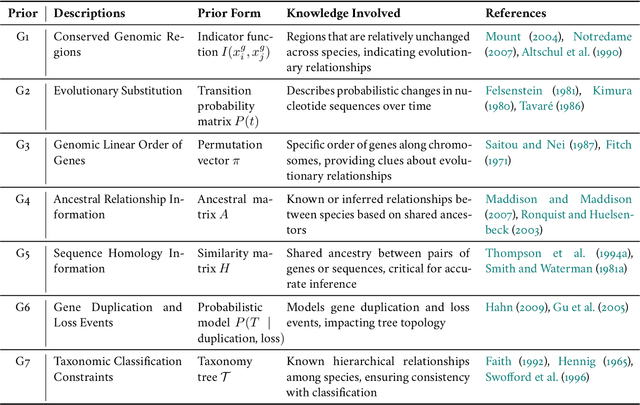

Abstract:Biological tree analysis serves as a pivotal tool in uncovering the evolutionary and differentiation relationships among organisms, genes, and cells. Its applications span diverse fields including phylogenetics, developmental biology, ecology, and medicine. Traditional tree inference methods, while foundational in early studies, face increasing limitations in processing the large-scale, complex datasets generated by modern high-throughput technologies. Recent advances in deep learning offer promising solutions, providing enhanced data processing and pattern recognition capabilities. However, challenges remain, particularly in accurately representing the inherently discrete and non-Euclidean nature of biological trees. In this review, we first outline the key biological priors fundamental to phylogenetic and differentiation tree analyses, facilitating a deeper interdisciplinary understanding between deep learning researchers and biologists. We then systematically examine the commonly used data formats and databases, serving as a comprehensive resource for model testing and development. We provide a critical analysis of traditional tree generation methods, exploring their underlying biological assumptions, technical characteristics, and limitations. Current developments in deep learning-based tree generation are reviewed, highlighting both recent advancements and existing challenges. Furthermore, we discuss the diverse applications of biological trees across various biological domains. Finally, we propose potential future directions and trends in leveraging deep learning for biological tree research, aiming to guide further exploration and innovation in this field.
Gentle-CLIP: Exploring Aligned Semantic In Low-Quality Multimodal Data With Soft Alignment
Jun 09, 2024Abstract:Multimodal fusion breaks through the barriers between diverse modalities and has already yielded numerous impressive performances. However, in various specialized fields, it is struggling to obtain sufficient alignment data for the training process, which seriously limits the use of previously elegant models. Thus, semi-supervised learning attempts to achieve multimodal alignment with fewer matched pairs but traditional methods like pseudo-labeling are difficult to apply in domains with no label information. To address these problems, we transform semi-supervised multimodal alignment into a manifold matching problem and propose a new method based on CLIP, named Gentle-CLIP. Specifically, we design a novel semantic density distribution loss to explore implicit semantic alignment information from unpaired multimodal data by constraining the latent representation distribution with fine granularity, thus eliminating the need for numerous strictly matched pairs. Meanwhile, we introduce multi-kernel maximum mean discrepancy as well as self-supervised contrastive loss to pull separate modality distributions closer and enhance the stability of the representation distribution. In addition, the contrastive loss used in CLIP is employed on the supervised matched data to prevent negative optimization. Extensive experiments conducted on a range of tasks in various fields, including protein, remote sensing, and the general vision-language field, demonstrate the effectiveness of our proposed Gentle-CLIP.
GenBench: A Benchmarking Suite for Systematic Evaluation of Genomic Foundation Models
Jun 01, 2024



Abstract:The Genomic Foundation Model (GFM) paradigm is expected to facilitate the extraction of generalizable representations from massive genomic data, thereby enabling their application across a spectrum of downstream applications. Despite advancements, a lack of evaluation framework makes it difficult to ensure equitable assessment due to experimental settings, model intricacy, benchmark datasets, and reproducibility challenges. In the absence of standardization, comparative analyses risk becoming biased and unreliable. To surmount this impasse, we introduce GenBench, a comprehensive benchmarking suite specifically tailored for evaluating the efficacy of Genomic Foundation Models. GenBench offers a modular and expandable framework that encapsulates a variety of state-of-the-art methodologies. Through systematic evaluations of datasets spanning diverse biological domains with a particular emphasis on both short-range and long-range genomic tasks, firstly including the three most important DNA tasks covering Coding Region, Non-Coding Region, Genome Structure, etc. Moreover, We provide a nuanced analysis of the interplay between model architecture and dataset characteristics on task-specific performance. Our findings reveal an interesting observation: independent of the number of parameters, the discernible difference in preference between the attention-based and convolution-based models on short- and long-range tasks may provide insights into the future design of GFM.
USD: Unsupervised Soft Contrastive Learning for Fault Detection in Multivariate Time Series
May 25, 2024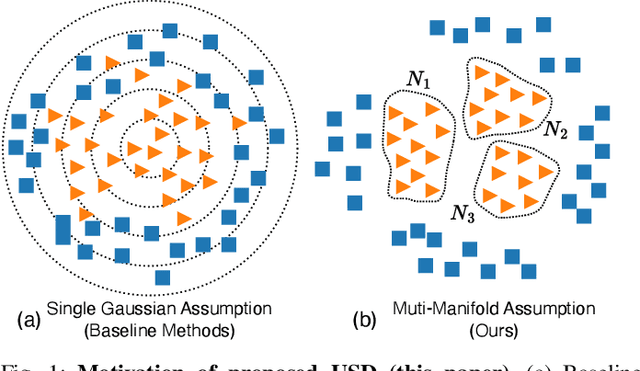
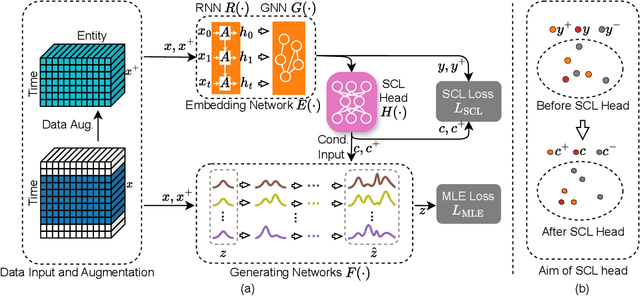
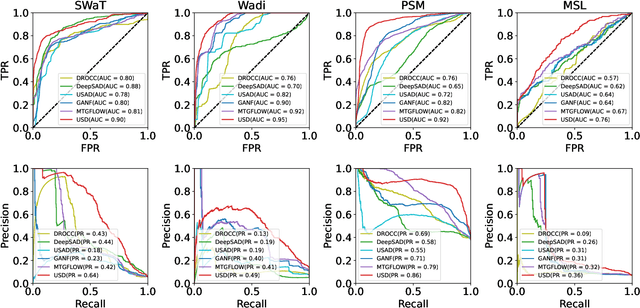
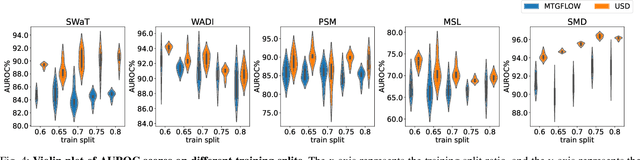
Abstract:Unsupervised fault detection in multivariate time series is critical for maintaining the integrity and efficiency of complex systems, with current methodologies largely focusing on statistical and machine learning techniques. However, these approaches often rest on the assumption that data distributions conform to Gaussian models, overlooking the diversity of patterns that can manifest in both normal and abnormal states, thereby diminishing discriminative performance. Our innovation addresses this limitation by introducing a combination of data augmentation and soft contrastive learning, specifically designed to capture the multifaceted nature of state behaviors more accurately. The data augmentation process enriches the dataset with varied representations of normal states, while soft contrastive learning fine-tunes the model's sensitivity to the subtle differences between normal and abnormal patterns, enabling it to recognize a broader spectrum of anomalies. This dual strategy significantly boosts the model's ability to distinguish between normal and abnormal states, leading to a marked improvement in fault detection performance across multiple datasets and settings, thereby setting a new benchmark for unsupervised fault detection in complex systems. The code of our method is available at \url{https://github.com/zangzelin/code_USD.git}.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge