Hang He
VULCA-Bench: A Multicultural Vision-Language Benchmark for Evaluating Cultural Understanding
Jan 12, 2026Abstract:We introduce VULCA-Bench, a multicultural art-critique benchmark for evaluating Vision-Language Models' (VLMs) cultural understanding beyond surface-level visual perception. Existing VLM benchmarks predominantly measure L1-L2 capabilities (object recognition, scene description, and factual question answering) while under-evaluate higher-order cultural interpretation. VULCA-Bench contains 7,410 matched image-critique pairs spanning eight cultural traditions, with Chinese-English bilingual coverage. We operationalise cultural understanding using a five-layer framework (L1-L5, from Visual Perception to Philosophical Aesthetics), instantiated as 225 culture-specific dimensions and supported by expert-written bilingual critiques. Our pilot results indicate that higher-layer reasoning (L3-L5) is consistently more challenging than visual and technical analysis (L1-L2). The dataset, evaluation scripts, and annotation tools are available under CC BY 4.0 in the supplementary materials.
ToolForge: A Data Synthesis Pipeline for Multi-Hop Search without Real-World APIs
Dec 18, 2025Abstract:Training LLMs to invoke tools and leverage retrieved information necessitates high-quality, diverse data. However, existing pipelines for synthetic data generation often rely on tens of thousands of real API calls to enhance generalization, incurring prohibitive costs while lacking multi-hop reasoning and self-reflection. To address these limitations, we introduce ToolForge, an automated synthesis framework that achieves strong real-world tool-calling performance by constructing only a small number of virtual tools, eliminating the need for real API calls. ToolForge leverages a (question, golden context, answer) triple to synthesize large-scale tool-learning data specifically designed for multi-hop search scenarios, further enriching the generated data through multi-hop reasoning and self-reflection mechanisms. To ensure data fidelity, we employ a Multi-Layer Validation Framework that integrates both rule-based and model-based assessments. Empirical results show that a model with only 8B parameters, when trained on our synthesized data, outperforms GPT-4o on multiple benchmarks. Our code and dataset are publicly available at https://github.com/Buycar-arb/ToolForge .
LocalSearchBench: Benchmarking Agentic Search in Real-World Local Life Services
Dec 08, 2025Abstract:Recent advances in large reasoning models (LRMs) have enabled agentic search systems to perform complex multi-step reasoning across multiple sources. However, most studies focus on general information retrieval and rarely explores vertical domains with unique challenges. In this work, we focus on local life services and introduce LocalSearchBench, which encompass diverse and complex business scenarios. Real-world queries in this domain are often ambiguous and require multi-hop reasoning across merchants and products, remaining challenging and not fully addressed. As the first comprehensive benchmark for agentic search in local life services, LocalSearchBench includes over 150,000 high-quality entries from various cities and business types. We construct 300 multi-hop QA tasks based on real user queries, challenging agents to understand questions and retrieve information in multiple steps. We also developed LocalPlayground, a unified environment integrating multiple tools for agent interaction. Experiments show that even state-of-the-art LRMs struggle on LocalSearchBench: the best model (DeepSeek-V3.1) achieves only 34.34% correctness, and most models have issues with completeness (average 77.33%) and faithfulness (average 61.99%). This highlights the need for specialized benchmarks and domain-specific agent training in local life services. Code, Benchmark, and Leaderboard are available at localsearchbench.github.io.
Promoting Efficient Reasoning with Verifiable Stepwise Reward
Aug 14, 2025Abstract:Large reasoning models (LRMs) have recently achieved significant progress in complex reasoning tasks, aided by reinforcement learning with verifiable rewards. However, LRMs often suffer from overthinking, expending excessive computation on simple problems and reducing efficiency. Existing efficient reasoning methods typically require accurate task assessment to preset token budgets or select reasoning modes, which limits their flexibility and reliability. In this work, we revisit the essence of overthinking and identify that encouraging effective steps while penalizing ineffective ones is key to its solution. To this end, we propose a novel rule-based verifiable stepwise reward mechanism (VSRM), which assigns rewards based on the performance of intermediate states in the reasoning trajectory. This approach is intuitive and naturally fits the step-by-step nature of reasoning tasks. We conduct extensive experiments on standard mathematical reasoning benchmarks, including AIME24 and AIME25, by integrating VSRM with PPO and Reinforce++. Results show that our method achieves substantial output length reduction while maintaining original reasoning performance, striking an optimal balance between efficiency and accuracy. Further analysis of overthinking frequency and pass@k score before and after training demonstrates that our approach in deed effectively suppresses ineffective steps and encourages effective reasoning, fundamentally alleviating the overthinking problem. All code will be released upon acceptance.
Automated detection of atomicity violations in large-scale systems
Apr 01, 2025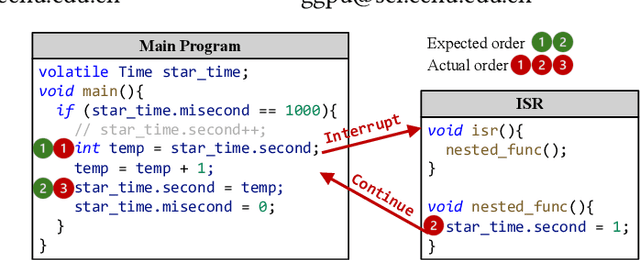

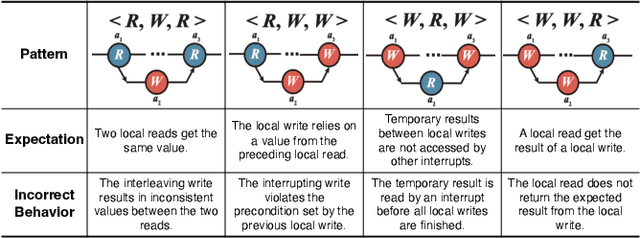

Abstract:Atomicity violations in interrupt-driven programs pose a significant threat to software safety in critical systems. These violations occur when the execution sequence of operations on shared resources is disrupted by asynchronous interrupts. Detecting atomicity violations is challenging due to the vast program state space, application-level code dependencies, and complex domain-specific knowledge. We propose Clover, a hybrid framework that integrates static analysis with large language model (LLM) agents to detect atomicity violations in real-world programs. Clover first performs static analysis to extract critical code snippets and operation information. It then initiates a multi-agent process, where the expert agent leverages domain-specific knowledge to detect atomicity violations, which are subsequently validated by the judge agent. Evaluations on RaceBench 2.1, SV-COMP, and RWIP demonstrate that Clover achieves a precision/recall of 92.3%/86.6%, outperforming existing approaches by 27.4-118.2% on F1-score.
FGBERT: Function-Driven Pre-trained Gene Language Model for Metagenomics
Feb 24, 2024



Abstract:Metagenomic data, comprising mixed multi-species genomes, are prevalent in diverse environments like oceans and soils, significantly impacting human health and ecological functions. However, current research relies on K-mer representations, limiting the capture of structurally relevant gene contexts. To address these limitations and further our understanding of complex relationships between metagenomic sequences and their functions, we introduce a protein-based gene representation as a context-aware and structure-relevant tokenizer. Our approach includes Masked Gene Modeling (MGM) for gene group-level pre-training, providing insights into inter-gene contextual information, and Triple Enhanced Metagenomic Contrastive Learning (TEM-CL) for gene-level pre-training to model gene sequence-function relationships. MGM and TEM-CL constitute our novel metagenomic language model {\NAME}, pre-trained on 100 million metagenomic sequences. We demonstrate the superiority of our proposed {\NAME} on eight datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge