Yucheng Guo
RL-VLA$^3$: Reinforcement Learning VLA Accelerating via Full Asynchronism
Feb 05, 2026Abstract:In recent years, Vision-Language-Action (VLA) models have emerged as a crucial pathway towards general embodied intelligence, yet their training efficiency has become a key bottleneck. Although existing reinforcement learning (RL)-based training frameworks like RLinf can enhance model generalization, they still rely on synchronous execution, leading to severe resource underutilization and throughput limitations during environment interaction, policy generation (rollout), and model update phases (actor). To overcome this challenge, this paper, for the first time, proposes and implements a fully-asynchronous policy training framework encompassing the entire pipeline from environment interaction, rollout generation, to actor policy updates. Systematically drawing inspiration from asynchronous optimization ideas in large model RL, our framework designs a multi-level decoupled architecture. This includes asynchronous parallelization of environment interaction and trajectory collection, streaming execution for policy generation, and decoupled scheduling for training updates. We validated the effectiveness of our method across diverse VLA models and environments. On the LIBERO benchmark, the framework achieves throughput improvements of up to 59.25\% compared to existing synchronous strategies. When deeply optimizing separation strategies, throughput can be increased by as much as 126.67\%. We verified the effectiveness of each asynchronous component via ablation studies. Scaling law validation across 8 to 256 GPUs demonstrates our method's excellent scalability under most conditions.
Time-series Forecast for Indoor Zone Air Temperature with Long Horizons: A Case Study with Sensor-based Data from a Smart Building
Dec 22, 2025Abstract:With the press of global climate change, extreme weather and sudden weather changes are becoming increasingly common. To maintain a comfortable indoor environment and minimize the contribution of the building to climate change as much as possible, higher requirements are placed on the operation and control of HVAC systems, e.g., more energy-efficient and flexible to response to the rapid change of weather. This places demands on the rapid modeling and prediction of zone air temperatures of buildings. Compared to the traditional simulation-based approach such as EnergyPlus and DOE2, a hybrid approach combined physics and data-driven is more suitable. Recently, the availability of high-quality datasets and algorithmic breakthroughs have driven a considerable amount of work in this field. However, in the niche of short- and long-term predictions, there are still some gaps in existing research. This paper aims to develop a time series forecast model to predict the zone air temperature in a building located in America on a 2-week horizon. The findings could be further improved to support intelligent control and operation of HVAC systems (i.e. demand flexibility) and could also be used as hybrid building energy modeling.
MergeDNA: Context-aware Genome Modeling with Dynamic Tokenization through Token Merging
Nov 17, 2025



Abstract:Modeling genomic sequences faces two unsolved challenges: the information density varies widely across different regions, while there is no clearly defined minimum vocabulary unit. Relying on either four primitive bases or independently designed DNA tokenizers, existing approaches with naive masked language modeling pre-training often fail to adapt to the varying complexities of genomic sequences. Leveraging Token Merging techniques, this paper introduces a hierarchical architecture that jointly optimizes a dynamic genomic tokenizer and latent Transformers with context-aware pre-training tasks. As for network structures, the tokenization module automatically chunks adjacent bases into words by stacking multiple layers of the differentiable token merging blocks with local-window constraints, then a Latent Encoder captures the global context of these merged words by full-attention blocks. Symmetrically employing a Latent Decoder and a Local Decoder, MergeDNA learns with two pre-training tasks: Merged Token Reconstruction simultaneously trains the dynamic tokenization module and adaptively filters important tokens, while Adaptive Masked Token Modeling learns to predict these filtered tokens to capture informative contents. Extensive experiments show that MergeDNA achieves superior performance on three popular DNA benchmarks and several multi-omics tasks with fine-tuning or zero-shot evaluation, outperforming typical tokenization methods and large-scale DNA foundation models.
Fine-Tuning Diffusion Generative Models via Rich Preference Optimization
Mar 13, 2025
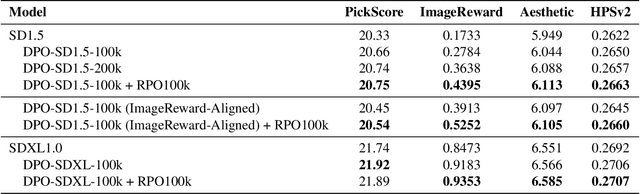
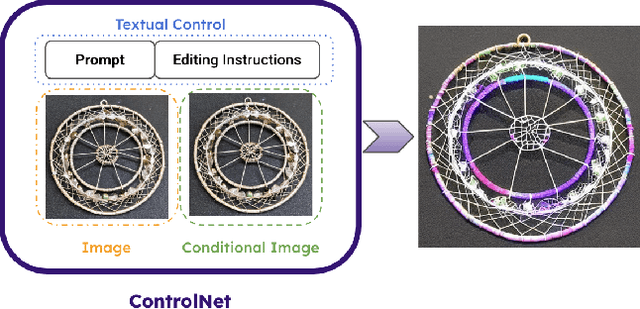
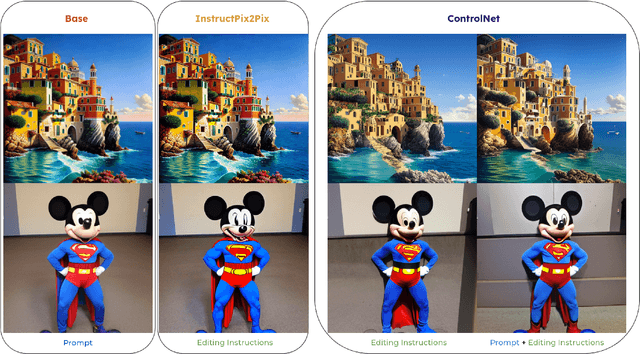
Abstract:We introduce Rich Preference Optimization (RPO), a novel pipeline that leverages rich feedback signals to improve the curation of preference pairs for fine-tuning text-to-image diffusion models. Traditional methods, like Diffusion-DPO, often rely solely on reward model labeling, which can be opaque, offer limited insights into the rationale behind preferences, and are prone to issues such as reward hacking or overfitting. In contrast, our approach begins with generating detailed critiques of synthesized images to extract reliable and actionable image editing instructions. By implementing these instructions, we create refined images, resulting in synthetic, informative preference pairs that serve as enhanced tuning datasets. We demonstrate the effectiveness of our pipeline and the resulting datasets in fine-tuning state-of-the-art diffusion models.
Life-Code: Central Dogma Modeling with Multi-Omics Sequence Unification
Feb 11, 2025



Abstract:The interactions between DNA, RNA, and proteins are fundamental to biological processes, as illustrated by the central dogma of molecular biology. While modern biological pre-trained models have achieved great success in analyzing these macromolecules individually, their interconnected nature remains under-explored. In this paper, we follow the guidance of the central dogma to redesign both the data and model pipeline and offer a comprehensive framework, Life-Code, that spans different biological functions. As for data flow, we propose a unified pipeline to integrate multi-omics data by reverse-transcribing RNA and reverse-translating amino acids into nucleotide-based sequences. As for the model, we design a codon tokenizer and a hybrid long-sequence architecture to encode the interactions of both coding and non-coding regions with masked modeling pre-training. To model the translation and folding process with coding sequences, Life-Code learns protein structures of the corresponding amino acids by knowledge distillation from off-the-shelf protein language models. Such designs enable Life-Code to capture complex interactions within genetic sequences, providing a more comprehensive understanding of multi-omics with the central dogma. Extensive Experiments show that Life-Code achieves state-of-the-art performance on various tasks across three omics, highlighting its potential for advancing multi-omics analysis and interpretation.
Drug Synergistic Combinations Predictions via Large-Scale Pre-Training and Graph Structure Learning
Jan 14, 2023



Abstract:Drug combination therapy is a well-established strategy for disease treatment with better effectiveness and less safety degradation. However, identifying novel drug combinations through wet-lab experiments is resource intensive due to the vast combinatorial search space. Recently, computational approaches, specifically deep learning models have emerged as an efficient way to discover synergistic combinations. While previous methods reported fair performance, their models usually do not take advantage of multi-modal data and they are unable to handle new drugs or cell lines. In this study, we collected data from various datasets covering various drug-related aspects. Then, we take advantage of large-scale pre-training models to generate informative representations and features for drugs, proteins, and diseases. Based on that, a message-passing graph is built on top to propagate information together with graph structure learning flexibility. This is first introduced in the biological networks and enables us to generate pseudo-relations in the graph. Our framework achieves state-of-the-art results in comparison with other deep learning-based methods on synergistic prediction benchmark datasets. We are also capable of inferencing new drug combination data in a test on an independent set released by AstraZeneca, where 10% of improvement over previous methods is observed. In addition, we're robust against unseen drugs and surpass almost 15% AU ROC compared to the second-best model. We believe our framework contributes to both the future wet-lab discovery of novel drugs and the building of promising guidance for precise combination medicine.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge