Vijay Prakash Dwivedi
PluRel: Synthetic Data unlocks Scaling Laws for Relational Foundation Models
Feb 03, 2026Abstract:Relational Foundation Models (RFMs) facilitate data-driven decision-making by learning from complex multi-table databases. However, the diverse relational databases needed to train such models are rarely public due to privacy constraints. While there are methods to generate synthetic tabular data of arbitrary size, incorporating schema structure and primary--foreign key connectivity for multi-table generation remains challenging. Here we introduce PluRel, a framework to synthesize multi-tabular relational databases from scratch. In a step-by-step fashion, PluRel models (1) schemas with directed graphs, (2) inter-table primary-foreign key connectivity with bipartite graphs, and, (3) feature distributions in tables via conditional causal mechanisms. The design space across these stages supports the synthesis of a wide range of diverse databases, while being computationally lightweight. Using PluRel, we observe for the first time that (1) RFM pretraining loss exhibits power-law scaling with the number of synthetic databases and total pretraining tokens, (2) scaling the number of synthetic databases improves generalization to real databases, and (3) synthetic pretraining yields strong base models for continued pretraining on real databases. Overall, our framework and results position synthetic data scaling as a promising paradigm for RFMs.
Large Language Models are Good Relational Learners
Jun 06, 2025



Abstract:Large language models (LLMs) have demonstrated remarkable capabilities across various domains, yet their application to relational deep learning (RDL) remains underexplored. Existing approaches adapt LLMs by traversing relational links between entities in a database and converting the structured data into flat text documents. Still, this text-based serialization disregards critical relational structures, introduces redundancy, and often exceeds standard LLM context lengths. We introduce Rel-LLM, a novel architecture that utilizes a graph neural network (GNN)- based encoder to generate structured relational prompts for LLMs within a retrieval-augmented generation (RAG) framework. Unlike traditional text-based serialization approaches, our method preserves the inherent relational structure of databases while enabling LLMs to effectively process and reason over complex entity relationships. Specifically, the GNN encoder extracts a local subgraph around an entity to build feature representations that contain relevant entity relationships and temporal dependencies. These representations are transformed into structured prompts using a denormalization process, effectively allowing the LLM to reason over relational structures. Through extensive experiments, we demonstrate that Rel-LLM outperforms existing methods on key RDL tasks, offering a scalable and efficient approach to integrating LLMs with structured data sources. Code is available at https://github.com/smiles724/Rel-LLM.
Relational Graph Transformer
May 16, 2025Abstract:Relational Deep Learning (RDL) is a promising approach for building state-of-the-art predictive models on multi-table relational data by representing it as a heterogeneous temporal graph. However, commonly used Graph Neural Network models suffer from fundamental limitations in capturing complex structural patterns and long-range dependencies that are inherent in relational data. While Graph Transformers have emerged as powerful alternatives to GNNs on general graphs, applying them to relational entity graphs presents unique challenges: (i) Traditional positional encodings fail to generalize to massive, heterogeneous graphs; (ii) existing architectures cannot model the temporal dynamics and schema constraints of relational data; (iii) existing tokenization schemes lose critical structural information. Here we introduce the Relational Graph Transformer (RelGT), the first graph transformer architecture designed specifically for relational tables. RelGT employs a novel multi-element tokenization strategy that decomposes each node into five components (features, type, hop distance, time, and local structure), enabling efficient encoding of heterogeneity, temporality, and topology without expensive precomputation. Our architecture combines local attention over sampled subgraphs with global attention to learnable centroids, incorporating both local and database-wide representations. Across 21 tasks from the RelBench benchmark, RelGT consistently matches or outperforms GNN baselines by up to 18%, establishing Graph Transformers as a powerful architecture for Relational Deep Learning.
Representation Learning of Structured Data for Medical Foundation Models
Oct 17, 2024

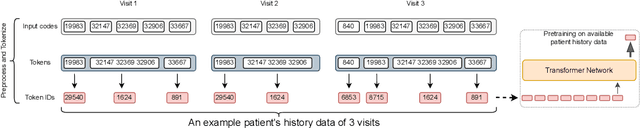
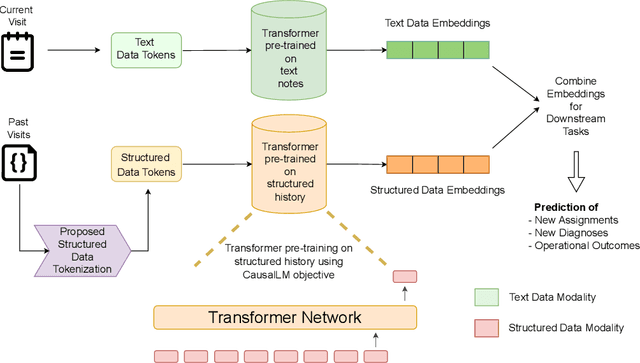
Abstract:Large Language Models (LLMs) have demonstrated remarkable performance across various domains, including healthcare. However, their ability to effectively represent structured non-textual data, such as the alphanumeric medical codes used in records like ICD-10 or SNOMED-CT, is limited and has been particularly exposed in recent research. This paper examines the challenges LLMs face in processing medical codes due to the shortcomings of current tokenization methods. As a result, we introduce the UniStruct architecture to design a multimodal medical foundation model of unstructured text and structured data, which addresses these challenges by adapting subword tokenization techniques specifically for the structured medical codes. Our approach is validated through model pre-training on both an extensive internal medical database and a public repository of structured medical records. Trained on over 1 billion tokens on the internal medical database, the proposed model achieves up to a 23% improvement in evaluation metrics, with around 2% gain attributed to our proposed tokenization. Additionally, when evaluated on the EHRSHOT public benchmark with a 1/1000 fraction of the pre-training data, the UniStruct model improves performance on over 42% of the downstream tasks. Our approach not only enhances the representation and generalization capabilities of patient-centric models but also bridges a critical gap in representation learning models' ability to handle complex structured medical data, alongside unstructured text.
MEDSAGE: Enhancing Robustness of Medical Dialogue Summarization to ASR Errors with LLM-generated Synthetic Dialogues
Aug 26, 2024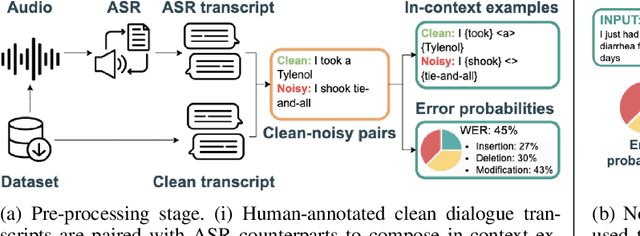
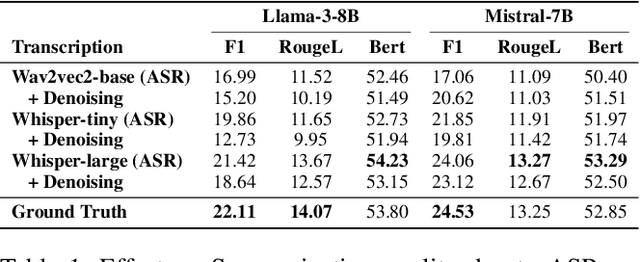

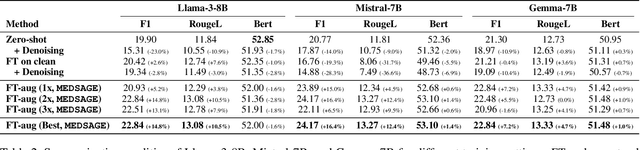
Abstract:Automatic Speech Recognition (ASR) systems are pivotal in transcribing speech into text, yet the errors they introduce can significantly degrade the performance of downstream tasks like summarization. This issue is particularly pronounced in clinical dialogue summarization, a low-resource domain where supervised data for fine-tuning is scarce, necessitating the use of ASR models as black-box solutions. Employing conventional data augmentation for enhancing the noise robustness of summarization models is not feasible either due to the unavailability of sufficient medical dialogue audio recordings and corresponding ASR transcripts. To address this challenge, we propose MEDSAGE, an approach for generating synthetic samples for data augmentation using Large Language Models (LLMs). Specifically, we leverage the in-context learning capabilities of LLMs and instruct them to generate ASR-like errors based on a few available medical dialogue examples with audio recordings. Experimental results show that LLMs can effectively model ASR noise, and incorporating this noisy data into the training process significantly improves the robustness and accuracy of medical dialogue summarization systems. This approach addresses the challenges of noisy ASR outputs in critical applications, offering a robust solution to enhance the reliability of clinical dialogue summarization.
uMedSum: A Unified Framework for Advancing Medical Abstractive Summarization
Aug 22, 2024



Abstract:Medical abstractive summarization faces the challenge of balancing faithfulness and informativeness. Current methods often sacrifice key information for faithfulness or introduce confabulations when prioritizing informativeness. While recent advancements in techniques like in-context learning (ICL) and fine-tuning have improved medical summarization, they often overlook crucial aspects such as faithfulness and informativeness without considering advanced methods like model reasoning and self-improvement. Moreover, the field lacks a unified benchmark, hindering systematic evaluation due to varied metrics and datasets. This paper addresses these gaps by presenting a comprehensive benchmark of six advanced abstractive summarization methods across three diverse datasets using five standardized metrics. Building on these findings, we propose uMedSum, a modular hybrid summarization framework that introduces novel approaches for sequential confabulation removal followed by key missing information addition, ensuring both faithfulness and informativeness. Our work improves upon previous GPT-4-based state-of-the-art (SOTA) medical summarization methods, significantly outperforming them in both quantitative metrics and qualitative domain expert evaluations. Notably, we achieve an average relative performance improvement of 11.8% in reference-free metrics over the previous SOTA. Doctors prefer uMedSum's summaries 6 times more than previous SOTA in difficult cases where there are chances of confabulations or missing information. These results highlight uMedSum's effectiveness and generalizability across various datasets and metrics, marking a significant advancement in medical summarization.
M-QALM: A Benchmark to Assess Clinical Reading Comprehension and Knowledge Recall in Large Language Models via Question Answering
Jun 06, 2024

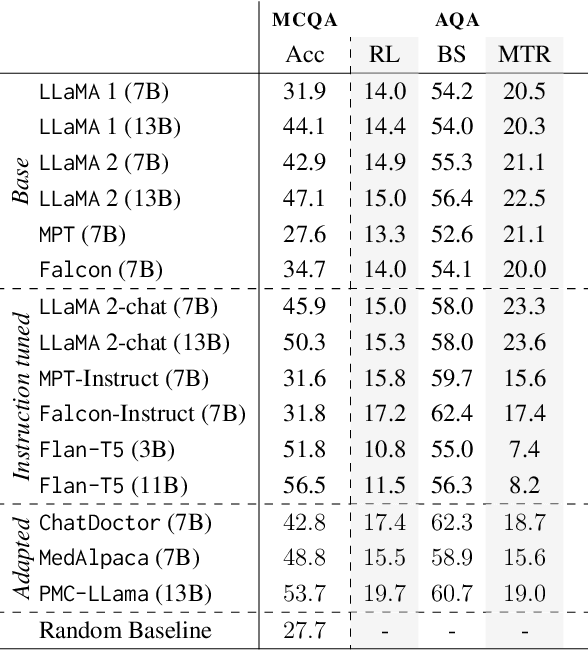

Abstract:There is vivid research on adapting Large Language Models (LLMs) to perform a variety of tasks in high-stakes domains such as healthcare. Despite their popularity, there is a lack of understanding of the extent and contributing factors that allow LLMs to recall relevant knowledge and combine it with presented information in the clinical and biomedical domain: a fundamental pre-requisite for success on down-stream tasks. Addressing this gap, we use Multiple Choice and Abstractive Question Answering to conduct a large-scale empirical study on 22 datasets in three generalist and three specialist biomedical sub-domains. Our multifaceted analysis of the performance of 15 LLMs, further broken down by sub-domain, source of knowledge and model architecture, uncovers success factors such as instruction tuning that lead to improved recall and comprehension. We further show that while recently proposed domain-adapted models may lack adequate knowledge, directly fine-tuning on our collected medical knowledge datasets shows encouraging results, even generalising to unseen specialist sub-domains. We complement the quantitative results with a skill-oriented manual error analysis, which reveals a significant gap between the models' capabilities to simply recall necessary knowledge and to integrate it with the presented context. To foster research and collaboration in this field we share M-QALM, our resources, standardised methodology, and evaluation results, with the research community to facilitate further advancements in clinical knowledge representation learning within language models.
Automated Clinical Coding for Outpatient Departments
Dec 24, 2023Abstract:Computerised clinical coding approaches aim to automate the process of assigning a set of codes to medical records. While there is active research pushing the state of the art on clinical coding for hospitalized patients, the outpatient setting -- where doctors tend to non-hospitalised patients -- is overlooked. Although both settings can be formalised as a multi-label classification task, they present unique and distinct challenges, which raises the question of whether the success of inpatient clinical coding approaches translates to the outpatient setting. This paper is the first to investigate how well state-of-the-art deep learning-based clinical coding approaches work in the outpatient setting at hospital scale. To this end, we collect a large outpatient dataset comprising over 7 million notes documenting over half a million patients. We adapt four state-of-the-art clinical coding approaches to this setting and evaluate their potential to assist coders. We find evidence that clinical coding in outpatient settings can benefit from more innovations in popular inpatient coding benchmarks. A deeper analysis of the factors contributing to the success -- amount and form of data and choice of document representation -- reveals the presence of easy-to-solve examples, the coding of which can be completely automated with a low error rate.
Graph Transformers for Large Graphs
Dec 18, 2023Abstract:Transformers have recently emerged as powerful neural networks for graph learning, showcasing state-of-the-art performance on several graph property prediction tasks. However, these results have been limited to small-scale graphs, where the computational feasibility of the global attention mechanism is possible. The next goal is to scale up these architectures to handle very large graphs on the scale of millions or even billions of nodes. With large-scale graphs, global attention learning is proven impractical due to its quadratic complexity w.r.t. the number of nodes. On the other hand, neighborhood sampling techniques become essential to manage large graph sizes, yet finding the optimal trade-off between speed and accuracy with sampling techniques remains challenging. This work advances representation learning on single large-scale graphs with a focus on identifying model characteristics and critical design constraints for developing scalable graph transformer (GT) architectures. We argue such GT requires layers that can adeptly learn both local and global graph representations while swiftly sampling the graph topology. As such, a key innovation of this work lies in the creation of a fast neighborhood sampling technique coupled with a local attention mechanism that encompasses a 4-hop reception field, but achieved through just 2-hop operations. This local node embedding is then integrated with a global node embedding, acquired via another self-attention layer with an approximate global codebook, before finally sent through a downstream layer for node predictions. The proposed GT framework, named LargeGT, overcomes previous computational bottlenecks and is validated on three large-scale node classification benchmarks. We report a 3x speedup and 16.8% performance gain on ogbn-products and snap-patents, while we also scale LargeGT on ogbn-papers100M with a 5.9% performance improvement.
Union Subgraph Neural Networks
May 25, 2023Abstract:Graph Neural Networks (GNNs) are widely used for graph representation learning in many application domains. The expressiveness of vanilla GNNs is upper-bounded by 1-dimensional Weisfeiler-Leman (1-WL) test as they operate on rooted subtrees through iterative message passing. In this paper, we empower GNNs by injecting neighbor-connectivity information extracted from a new type of substructure. We first investigate different kinds of connectivities existing in a local neighborhood and identify a substructure called union subgraph, which is able to capture the complete picture of the 1-hop neighborhood of an edge. We then design a shortest-path-based substructure descriptor that possesses three nice properties and can effectively encode the high-order connectivities in union subgraphs. By infusing the encoded neighbor connectivities, we propose a novel model, namely Union Subgraph Neural Network (UnionSNN), which is proven to be strictly more powerful than 1-WL in distinguishing non-isomorphic graphs. Additionally, the local encoding from union subgraphs can also be injected into arbitrary message-passing neural networks (MPNNs) and Transformer-based models as a plugin. Extensive experiments on 17 benchmarks of both graph-level and node-level tasks demonstrate that UnionSNN outperforms state-of-the-art baseline models, with competitive computational efficiency. The injection of our local encoding to existing models is able to boost the performance by up to 11.09%.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge