M-QALM: A Benchmark to Assess Clinical Reading Comprehension and Knowledge Recall in Large Language Models via Question Answering
Paper and Code
Jun 06, 2024

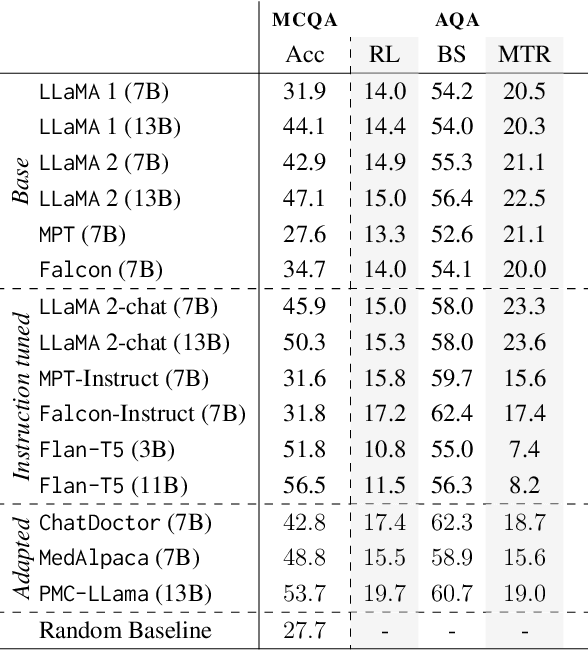

There is vivid research on adapting Large Language Models (LLMs) to perform a variety of tasks in high-stakes domains such as healthcare. Despite their popularity, there is a lack of understanding of the extent and contributing factors that allow LLMs to recall relevant knowledge and combine it with presented information in the clinical and biomedical domain: a fundamental pre-requisite for success on down-stream tasks. Addressing this gap, we use Multiple Choice and Abstractive Question Answering to conduct a large-scale empirical study on 22 datasets in three generalist and three specialist biomedical sub-domains. Our multifaceted analysis of the performance of 15 LLMs, further broken down by sub-domain, source of knowledge and model architecture, uncovers success factors such as instruction tuning that lead to improved recall and comprehension. We further show that while recently proposed domain-adapted models may lack adequate knowledge, directly fine-tuning on our collected medical knowledge datasets shows encouraging results, even generalising to unseen specialist sub-domains. We complement the quantitative results with a skill-oriented manual error analysis, which reveals a significant gap between the models' capabilities to simply recall necessary knowledge and to integrate it with the presented context. To foster research and collaboration in this field we share M-QALM, our resources, standardised methodology, and evaluation results, with the research community to facilitate further advancements in clinical knowledge representation learning within language models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge