Stefan Winkler
Representation Learning of Structured Data for Medical Foundation Models
Oct 17, 2024

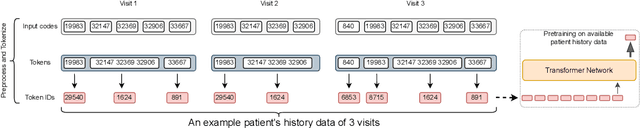
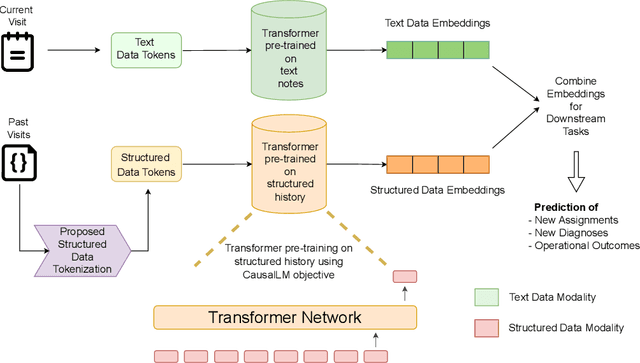
Abstract:Large Language Models (LLMs) have demonstrated remarkable performance across various domains, including healthcare. However, their ability to effectively represent structured non-textual data, such as the alphanumeric medical codes used in records like ICD-10 or SNOMED-CT, is limited and has been particularly exposed in recent research. This paper examines the challenges LLMs face in processing medical codes due to the shortcomings of current tokenization methods. As a result, we introduce the UniStruct architecture to design a multimodal medical foundation model of unstructured text and structured data, which addresses these challenges by adapting subword tokenization techniques specifically for the structured medical codes. Our approach is validated through model pre-training on both an extensive internal medical database and a public repository of structured medical records. Trained on over 1 billion tokens on the internal medical database, the proposed model achieves up to a 23% improvement in evaluation metrics, with around 2% gain attributed to our proposed tokenization. Additionally, when evaluated on the EHRSHOT public benchmark with a 1/1000 fraction of the pre-training data, the UniStruct model improves performance on over 42% of the downstream tasks. Our approach not only enhances the representation and generalization capabilities of patient-centric models but also bridges a critical gap in representation learning models' ability to handle complex structured medical data, alongside unstructured text.
Seemingly Plausible Distractors in Multi-Hop Reasoning: Are Large Language Models Attentive Readers?
Sep 08, 2024Abstract:State-of-the-art Large Language Models (LLMs) are accredited with an increasing number of different capabilities, ranging from reading comprehension, over advanced mathematical and reasoning skills to possessing scientific knowledge. In this paper we focus on their multi-hop reasoning capability: the ability to identify and integrate information from multiple textual sources. Given the concerns with the presence of simplifying cues in existing multi-hop reasoning benchmarks, which allow models to circumvent the reasoning requirement, we set out to investigate, whether LLMs are prone to exploiting such simplifying cues. We find evidence that they indeed circumvent the requirement to perform multi-hop reasoning, but they do so in more subtle ways than what was reported about their fine-tuned pre-trained language model (PLM) predecessors. Motivated by this finding, we propose a challenging multi-hop reasoning benchmark, by generating seemingly plausible multi-hop reasoning chains, which ultimately lead to incorrect answers. We evaluate multiple open and proprietary state-of-the-art LLMs, and find that their performance to perform multi-hop reasoning is affected, as indicated by up to 45% relative decrease in F1 score when presented with such seemingly plausible alternatives. We conduct a deeper analysis and find evidence that while LLMs tend to ignore misleading lexical cues, misleading reasoning paths indeed present a significant challenge.
MEDSAGE: Enhancing Robustness of Medical Dialogue Summarization to ASR Errors with LLM-generated Synthetic Dialogues
Aug 26, 2024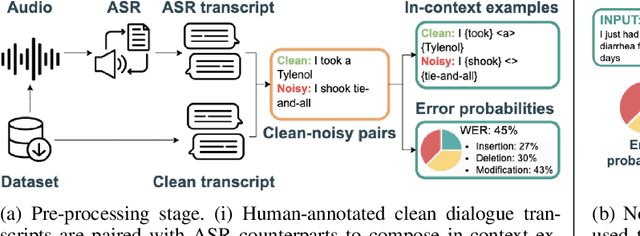
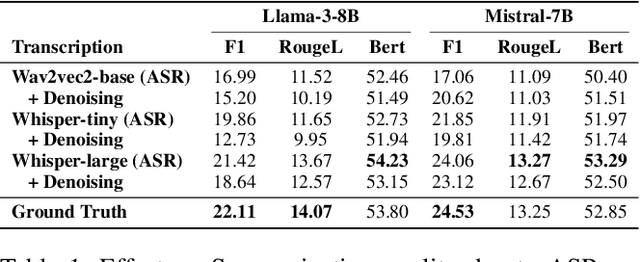

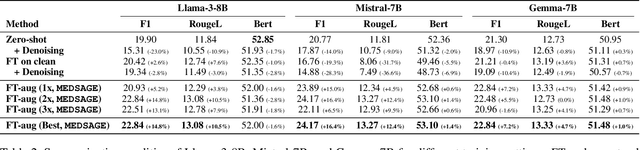
Abstract:Automatic Speech Recognition (ASR) systems are pivotal in transcribing speech into text, yet the errors they introduce can significantly degrade the performance of downstream tasks like summarization. This issue is particularly pronounced in clinical dialogue summarization, a low-resource domain where supervised data for fine-tuning is scarce, necessitating the use of ASR models as black-box solutions. Employing conventional data augmentation for enhancing the noise robustness of summarization models is not feasible either due to the unavailability of sufficient medical dialogue audio recordings and corresponding ASR transcripts. To address this challenge, we propose MEDSAGE, an approach for generating synthetic samples for data augmentation using Large Language Models (LLMs). Specifically, we leverage the in-context learning capabilities of LLMs and instruct them to generate ASR-like errors based on a few available medical dialogue examples with audio recordings. Experimental results show that LLMs can effectively model ASR noise, and incorporating this noisy data into the training process significantly improves the robustness and accuracy of medical dialogue summarization systems. This approach addresses the challenges of noisy ASR outputs in critical applications, offering a robust solution to enhance the reliability of clinical dialogue summarization.
LLMs are not Zero-Shot Reasoners for Biomedical Information Extraction
Aug 22, 2024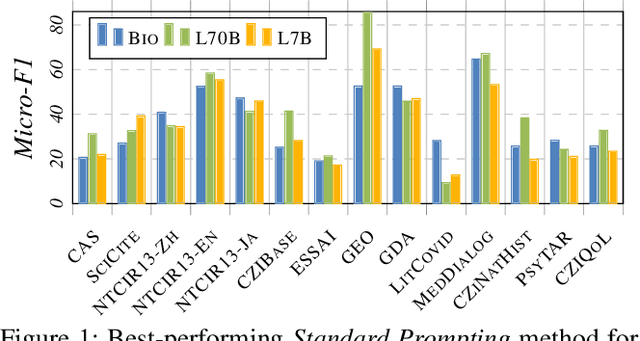
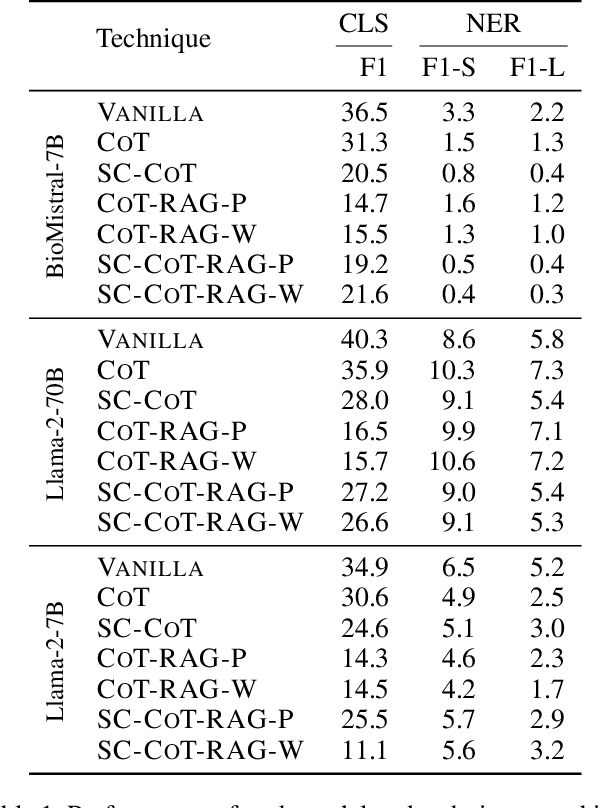
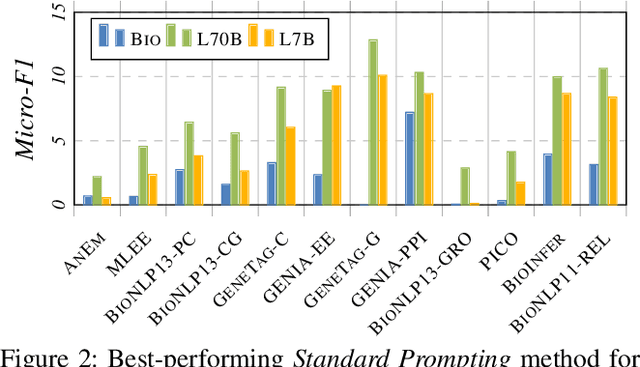
Abstract:Large Language Models (LLMs) are increasingly adopted for applications in healthcare, reaching the performance of domain experts on tasks such as question answering and document summarisation. Despite their success on these tasks, it is unclear how well LLMs perform on tasks that are traditionally pursued in the biomedical domain, such as structured information extration. To breach this gap, in this paper, we systematically benchmark LLM performance in Medical Classification and Named Entity Recognition (NER) tasks. We aim to disentangle the contribution of different factors to the performance, particularly the impact of LLMs' task knowledge and reasoning capabilities, their (parametric) domain knowledge, and addition of external knowledge. To this end we evaluate various open LLMs -- including BioMistral and Llama-2 models -- on a diverse set of biomedical datasets, using standard prompting, Chain-of-Thought (CoT) and Self-Consistency based reasoning as well as Retrieval-Augmented Generation (RAG) with PubMed and Wikipedia corpora. Counter-intuitively, our results reveal that standard prompting consistently outperforms more complex techniques across both tasks, laying bare the limitations in the current application of CoT, self-consistency and RAG in the biomedical domain. Our findings suggest that advanced prompting methods developed for knowledge- or reasoning-intensive tasks, such as CoT or RAG, are not easily portable to biomedical tasks where precise structured outputs are required. This highlights the need for more effective integration of external knowledge and reasoning mechanisms in LLMs to enhance their performance in real-world biomedical applications.
M-QALM: A Benchmark to Assess Clinical Reading Comprehension and Knowledge Recall in Large Language Models via Question Answering
Jun 06, 2024

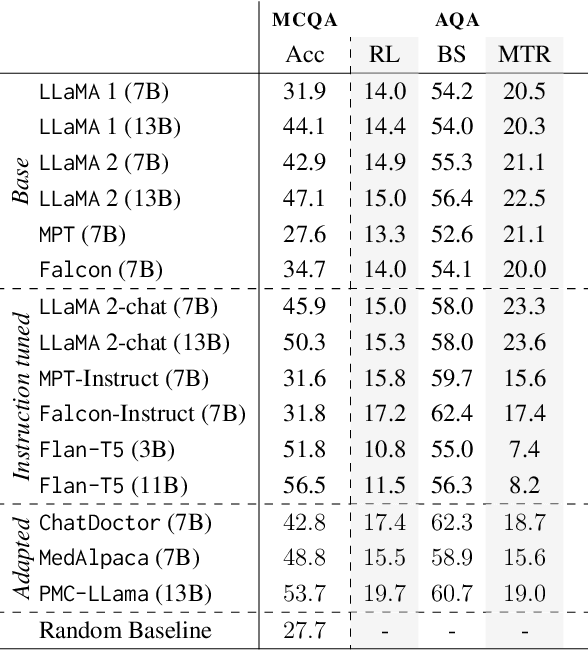

Abstract:There is vivid research on adapting Large Language Models (LLMs) to perform a variety of tasks in high-stakes domains such as healthcare. Despite their popularity, there is a lack of understanding of the extent and contributing factors that allow LLMs to recall relevant knowledge and combine it with presented information in the clinical and biomedical domain: a fundamental pre-requisite for success on down-stream tasks. Addressing this gap, we use Multiple Choice and Abstractive Question Answering to conduct a large-scale empirical study on 22 datasets in three generalist and three specialist biomedical sub-domains. Our multifaceted analysis of the performance of 15 LLMs, further broken down by sub-domain, source of knowledge and model architecture, uncovers success factors such as instruction tuning that lead to improved recall and comprehension. We further show that while recently proposed domain-adapted models may lack adequate knowledge, directly fine-tuning on our collected medical knowledge datasets shows encouraging results, even generalising to unseen specialist sub-domains. We complement the quantitative results with a skill-oriented manual error analysis, which reveals a significant gap between the models' capabilities to simply recall necessary knowledge and to integrate it with the presented context. To foster research and collaboration in this field we share M-QALM, our resources, standardised methodology, and evaluation results, with the research community to facilitate further advancements in clinical knowledge representation learning within language models.
Automated Clinical Coding for Outpatient Departments
Dec 24, 2023Abstract:Computerised clinical coding approaches aim to automate the process of assigning a set of codes to medical records. While there is active research pushing the state of the art on clinical coding for hospitalized patients, the outpatient setting -- where doctors tend to non-hospitalised patients -- is overlooked. Although both settings can be formalised as a multi-label classification task, they present unique and distinct challenges, which raises the question of whether the success of inpatient clinical coding approaches translates to the outpatient setting. This paper is the first to investigate how well state-of-the-art deep learning-based clinical coding approaches work in the outpatient setting at hospital scale. To this end, we collect a large outpatient dataset comprising over 7 million notes documenting over half a million patients. We adapt four state-of-the-art clinical coding approaches to this setting and evaluate their potential to assist coders. We find evidence that clinical coding in outpatient settings can benefit from more innovations in popular inpatient coding benchmarks. A deeper analysis of the factors contributing to the success -- amount and form of data and choice of document representation -- reveals the presence of easy-to-solve examples, the coding of which can be completely automated with a low error rate.
MaxEnt Loss: Constrained Maximum Entropy for Calibration under Out-of-Distribution Shift
Oct 26, 2023



Abstract:We present a new loss function that addresses the out-of-distribution (OOD) calibration problem. While many objective functions have been proposed to effectively calibrate models in-distribution, our findings show that they do not always fare well OOD. Based on the Principle of Maximum Entropy, we incorporate helpful statistical constraints observed during training, delivering better model calibration without sacrificing accuracy. We provide theoretical analysis and show empirically that our method works well in practice, achieving state-of-the-art calibration on both synthetic and real-world benchmarks.
PULSAR at MEDIQA-Sum 2023: Large Language Models Augmented by Synthetic Dialogue Convert Patient Dialogues to Medical Records
Jul 05, 2023


Abstract:This paper describes PULSAR, our system submission at the ImageClef 2023 MediQA-Sum task on summarising patient-doctor dialogues into clinical records. The proposed framework relies on domain-specific pre-training, to produce a specialised language model which is trained on task-specific natural data augmented by synthetic data generated by a black-box LLM. We find limited evidence towards the efficacy of domain-specific pre-training and data augmentation, while scaling up the language model yields the best performance gains. Our approach was ranked second and third among 13 submissions on task B of the challenge. Our code is available at https://github.com/yuping-wu/PULSAR.
PULSAR: Pre-training with Extracted Healthcare Terms for Summarising Patients' Problems and Data Augmentation with Black-box Large Language Models
Jun 05, 2023



Abstract:Medical progress notes play a crucial role in documenting a patient's hospital journey, including his or her condition, treatment plan, and any updates for healthcare providers. Automatic summarisation of a patient's problems in the form of a problem list can aid stakeholders in understanding a patient's condition, reducing workload and cognitive bias. BioNLP 2023 Shared Task 1A focuses on generating a list of diagnoses and problems from the provider's progress notes during hospitalisation. In this paper, we introduce our proposed approach to this task, which integrates two complementary components. One component employs large language models (LLMs) for data augmentation; the other is an abstractive summarisation LLM with a novel pre-training objective for generating the patients' problems summarised as a list. Our approach was ranked second among all submissions to the shared task. The performance of our model on the development and test datasets shows that our approach is more robust on unknown data, with an improvement of up to 3.1 points over the same size of the larger model.
A Two-Stage Decoder for Efficient ICD Coding
May 27, 2023



Abstract:Clinical notes in healthcare facilities are tagged with the International Classification of Diseases (ICD) code; a list of classification codes for medical diagnoses and procedures. ICD coding is a challenging multilabel text classification problem due to noisy clinical document inputs and long-tailed label distribution. Recent automated ICD coding efforts improve performance by encoding medical notes and codes with additional data and knowledge bases. However, most of them do not reflect how human coders generate the code: first, the coders select general code categories and then look for specific subcategories that are relevant to a patient's condition. Inspired by this, we propose a two-stage decoding mechanism to predict ICD codes. Our model uses the hierarchical properties of the codes to split the prediction into two steps: At first, we predict the parent code and then predict the child code based on the previous prediction. Experiments on the public MIMIC-III data set show that our model performs well in single-model settings without external data or knowledge.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge