Yuping Wu
SynBench: A Benchmark for Differentially Private Text Generation
Sep 18, 2025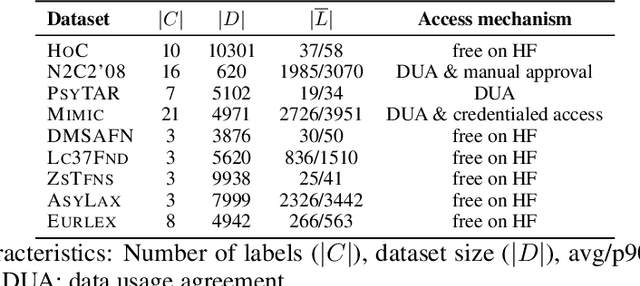
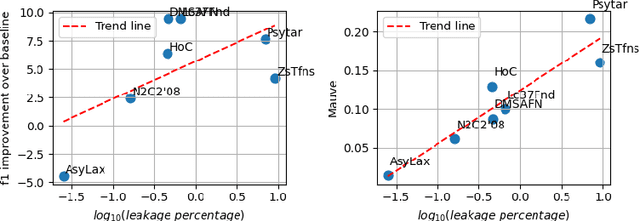
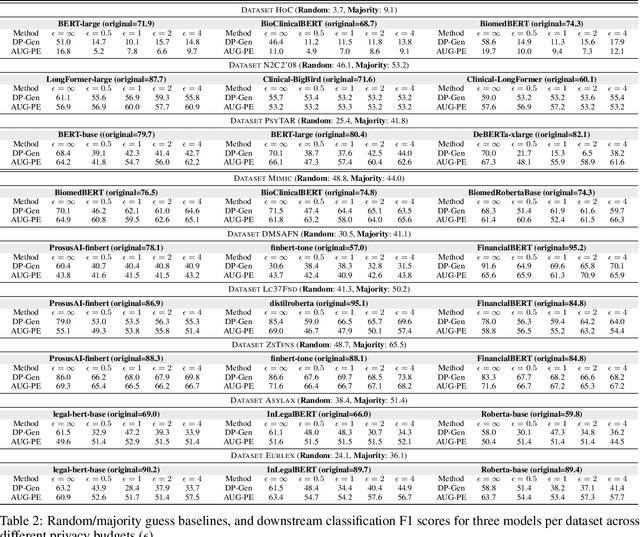
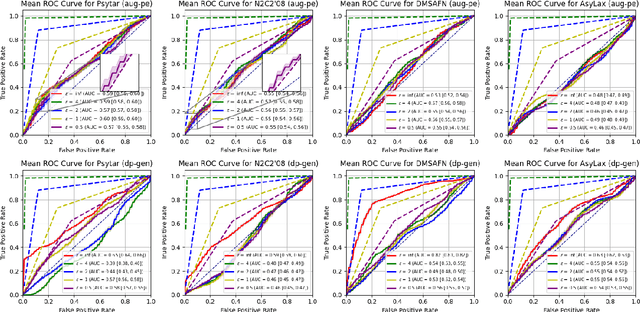
Abstract:Data-driven decision support in high-stakes domains like healthcare and finance faces significant barriers to data sharing due to regulatory, institutional, and privacy concerns. While recent generative AI models, such as large language models, have shown impressive performance in open-domain tasks, their adoption in sensitive environments remains limited by unpredictable behaviors and insufficient privacy-preserving datasets for benchmarking. Existing anonymization methods are often inadequate, especially for unstructured text, as redaction and masking can still allow re-identification. Differential Privacy (DP) offers a principled alternative, enabling the generation of synthetic data with formal privacy assurances. In this work, we address these challenges through three key contributions. First, we introduce a comprehensive evaluation framework with standardized utility and fidelity metrics, encompassing nine curated datasets that capture domain-specific complexities such as technical jargon, long-context dependencies, and specialized document structures. Second, we conduct a large-scale empirical study benchmarking state-of-the-art DP text generation methods and LLMs of varying sizes and different fine-tuning strategies, revealing that high-quality domain-specific synthetic data generation under DP constraints remains an unsolved challenge, with performance degrading as domain complexity increases. Third, we develop a membership inference attack (MIA) methodology tailored for synthetic text, providing first empirical evidence that the use of public datasets - potentially present in pre-training corpora - can invalidate claimed privacy guarantees. Our findings underscore the urgent need for rigorous privacy auditing and highlight persistent gaps between open-domain and specialist evaluations, informing responsible deployment of generative AI in privacy-sensitive, high-stakes settings.
Extract-and-Abstract: Unifying Extractive and Abstractive Summarization within Single Encoder-Decoder Framework
Sep 18, 2024



Abstract:Extract-then-Abstract is a naturally coherent paradigm to conduct abstractive summarization with the help of salient information identified by the extractive model. Previous works that adopt this paradigm train the extractor and abstractor separately and introduce extra parameters to highlight the extracted salients to the abstractor, which results in error accumulation and additional training costs. In this paper, we first introduce a parameter-free highlight method into the encoder-decoder framework: replacing the encoder attention mask with a saliency mask in the cross-attention module to force the decoder to focus only on salient parts of the input. A preliminary analysis compares different highlight methods, demonstrating the effectiveness of our saliency mask. We further propose the novel extract-and-abstract paradigm, ExtAbs, which jointly and seamlessly performs Extractive and Abstractive summarization tasks within single encoder-decoder model to reduce error accumulation. In ExtAbs, the vanilla encoder is augmented to extract salients, and the vanilla decoder is modified with the proposed saliency mask to generate summaries. Built upon BART and PEGASUS, experiments on three datasets show that ExtAbs can achieve superior performance than baselines on the extractive task and performs comparable, or even better than the vanilla models on the abstractive task.
LLMs are not Zero-Shot Reasoners for Biomedical Information Extraction
Aug 22, 2024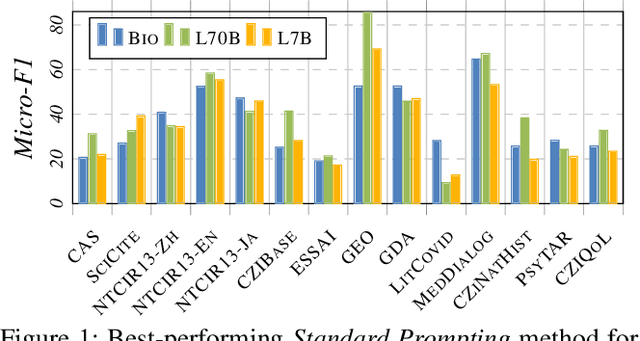
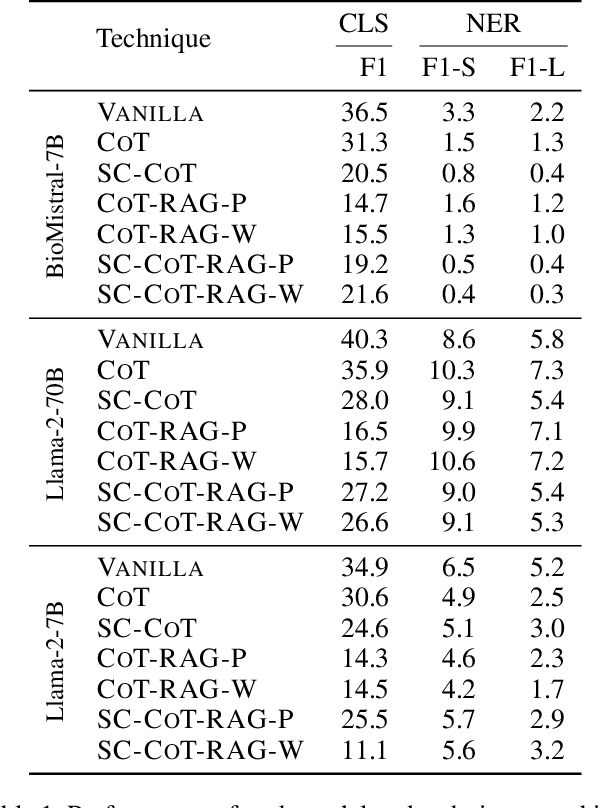
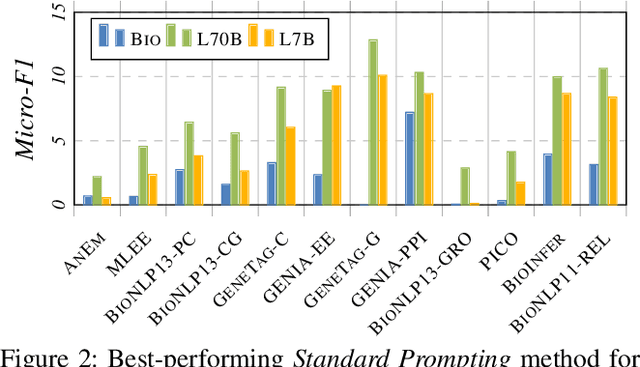
Abstract:Large Language Models (LLMs) are increasingly adopted for applications in healthcare, reaching the performance of domain experts on tasks such as question answering and document summarisation. Despite their success on these tasks, it is unclear how well LLMs perform on tasks that are traditionally pursued in the biomedical domain, such as structured information extration. To breach this gap, in this paper, we systematically benchmark LLM performance in Medical Classification and Named Entity Recognition (NER) tasks. We aim to disentangle the contribution of different factors to the performance, particularly the impact of LLMs' task knowledge and reasoning capabilities, their (parametric) domain knowledge, and addition of external knowledge. To this end we evaluate various open LLMs -- including BioMistral and Llama-2 models -- on a diverse set of biomedical datasets, using standard prompting, Chain-of-Thought (CoT) and Self-Consistency based reasoning as well as Retrieval-Augmented Generation (RAG) with PubMed and Wikipedia corpora. Counter-intuitively, our results reveal that standard prompting consistently outperforms more complex techniques across both tasks, laying bare the limitations in the current application of CoT, self-consistency and RAG in the biomedical domain. Our findings suggest that advanced prompting methods developed for knowledge- or reasoning-intensive tasks, such as CoT or RAG, are not easily portable to biomedical tasks where precise structured outputs are required. This highlights the need for more effective integration of external knowledge and reasoning mechanisms in LLMs to enhance their performance in real-world biomedical applications.
Which Side Are You On? A Multi-task Dataset for End-to-End Argument Summarisation and Evaluation
Jun 06, 2024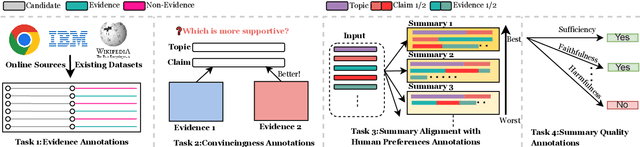
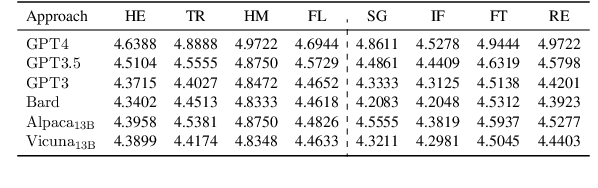
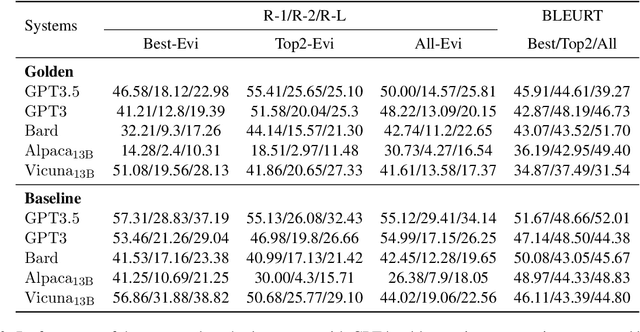
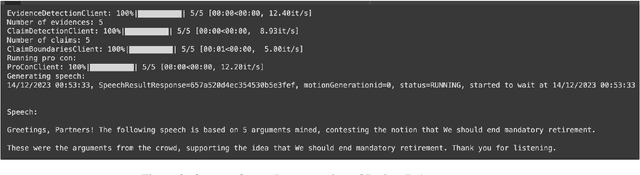
Abstract:With the recent advances of large language models (LLMs), it is no longer infeasible to build an automated debate system that helps people to synthesise persuasive arguments. Previous work attempted this task by integrating multiple components. In our work, we introduce an argument mining dataset that captures the end-to-end process of preparing an argumentative essay for a debate, which covers the tasks of claim and evidence identification (Task 1 ED), evidence convincingness ranking (Task 2 ECR), argumentative essay summarisation and human preference ranking (Task 3 ASR) and metric learning for automated evaluation of resulting essays, based on human feedback along argument quality dimensions (Task 4 SQE). Our dataset contains 14k examples of claims that are fully annotated with the various properties supporting the aforementioned tasks. We evaluate multiple generative baselines for each of these tasks, including representative LLMs. We find, that while they show promising results on individual tasks in our benchmark, their end-to-end performance on all four tasks in succession deteriorates significantly, both in automated measures as well as in human-centred evaluation. This challenge presented by our proposed dataset motivates future research on end-to-end argument mining and summarisation. The repository of this project is available at https://github.com/HarrywillDr/ArgSum-Datatset
PULSAR at MEDIQA-Sum 2023: Large Language Models Augmented by Synthetic Dialogue Convert Patient Dialogues to Medical Records
Jul 05, 2023


Abstract:This paper describes PULSAR, our system submission at the ImageClef 2023 MediQA-Sum task on summarising patient-doctor dialogues into clinical records. The proposed framework relies on domain-specific pre-training, to produce a specialised language model which is trained on task-specific natural data augmented by synthetic data generated by a black-box LLM. We find limited evidence towards the efficacy of domain-specific pre-training and data augmentation, while scaling up the language model yields the best performance gains. Our approach was ranked second and third among 13 submissions on task B of the challenge. Our code is available at https://github.com/yuping-wu/PULSAR.
PULSAR: Pre-training with Extracted Healthcare Terms for Summarising Patients' Problems and Data Augmentation with Black-box Large Language Models
Jun 05, 2023



Abstract:Medical progress notes play a crucial role in documenting a patient's hospital journey, including his or her condition, treatment plan, and any updates for healthcare providers. Automatic summarisation of a patient's problems in the form of a problem list can aid stakeholders in understanding a patient's condition, reducing workload and cognitive bias. BioNLP 2023 Shared Task 1A focuses on generating a list of diagnoses and problems from the provider's progress notes during hospitalisation. In this paper, we introduce our proposed approach to this task, which integrates two complementary components. One component employs large language models (LLMs) for data augmentation; the other is an abstractive summarisation LLM with a novel pre-training objective for generating the patients' problems summarised as a list. Our approach was ranked second among all submissions to the shared task. The performance of our model on the development and test datasets shows that our approach is more robust on unknown data, with an improvement of up to 3.1 points over the same size of the larger model.
On Cross-Domain Pre-Trained Language Models for Clinical Text Mining: How Do They Perform on Data-Constrained Fine-Tuning?
Oct 31, 2022Abstract:Pre-trained language models (PLMs) have been deployed in many natural language processing (NLP) tasks and in various domains. Language model pre-training from general or mixed domain rich data plus fine-tuning using small amounts of available data in a low resource domain demonstrated beneficial results by researchers. In this work, we question this statement and verify if BERT-based PLMs from the biomedical domain can perform well in clinical text mining tasks via fine-tuning. We test the state-of-the-art models, i.e. Bioformer which is pre-trained on a large amount of biomedical data from PubMed corpus. We use a historical n2c2 clinical NLP challenge dataset for fine-tuning its task-adapted version (BioformerApt), and show that their performances are actually very low. We also present our own end-to-end model, TransformerCRF, which is developed using Transformer and conditional random fields (CRFs) as encoder and decoder. We further create a new variation model by adding a CRF layer on top of PLM Bioformer (BioformerCRF). We investigate the performances of TransformerCRF on clinical text mining tasks by training from scratch using a limited amount of data, as well as the model BioformerCRF. Experimental evaluation shows that, in a \textit{constrained setting}, all tested models are \textit{far from ideal} regarding extreme low-frequency special token recognition, even though they can achieve relatively higher accuracy on overall text tagging. Our models including source codes will be hosted at \url{https://github.com/poethan/TransformerCRF}.
EDU-level Extractive Summarization with Varying Summary Lengths
Oct 08, 2022
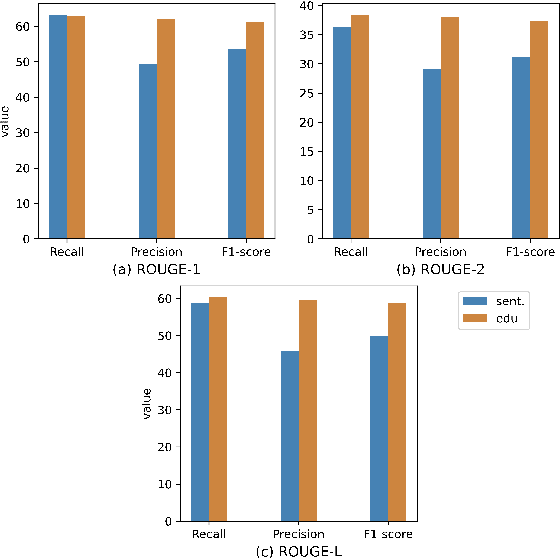
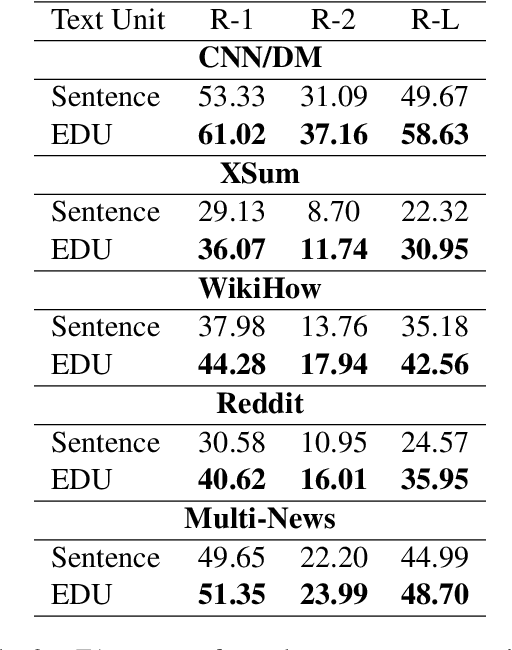
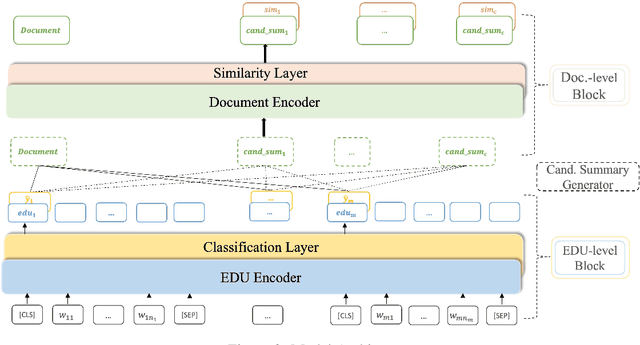
Abstract:Extractive models usually formulate text summarization as extracting top-k important sentences from document as summary. Few work exploited extracting finer-grained Elementary Discourse Unit (EDU) and there is little analysis and justification for the extractive unit selection. To fill such a gap, this paper firstly conducts oracle analysis to compare the upper bound of performance for models based on EDUs and sentences. The analysis provides evidences from both theoretical and experimental perspectives to justify that EDUs make more concise and precise summary than sentences without losing salient information. Then, considering this merit of EDUs, this paper further proposes EDU-level extractive model with Varying summary Lengths (EDU-VL) and develops the corresponding learning algorithm. EDU-VL learns to encode and predict probabilities of EDUs in document, and encode EDU-level candidate summaries with different lengths based on various $k$ values and select the best candidate summary in an end-to-end training manner. Finally, the proposed and developed approach is experimented on single and multi-document benchmark datasets and shows the improved performances in comparison with the state-of-the-art models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge