Aishik Nagar
Zero-Shot Visual Reasoning by Vision-Language Models: Benchmarking and Analysis
Aug 27, 2024



Abstract:Vision-language models (VLMs) have shown impressive zero- and few-shot performance on real-world visual question answering (VQA) benchmarks, alluding to their capabilities as visual reasoning engines. However, the benchmarks being used conflate "pure" visual reasoning with world knowledge, and also have questions that involve a limited number of reasoning steps. Thus, it remains unclear whether a VLM's apparent visual reasoning performance is due to its world knowledge, or due to actual visual reasoning capabilities. To clarify this ambiguity, we systematically benchmark and dissect the zero-shot visual reasoning capabilities of VLMs through synthetic datasets that require minimal world knowledge, and allow for analysis over a broad range of reasoning steps. We focus on two novel aspects of zero-shot visual reasoning: i) evaluating the impact of conveying scene information as either visual embeddings or purely textual scene descriptions to the underlying large language model (LLM) of the VLM, and ii) comparing the effectiveness of chain-of-thought prompting to standard prompting for zero-shot visual reasoning. We find that the underlying LLMs, when provided textual scene descriptions, consistently perform better compared to being provided visual embeddings. In particular, 18% higher accuracy is achieved on the PTR dataset. We also find that CoT prompting performs marginally better than standard prompting only for the comparatively large GPT-3.5-Turbo (175B) model, and does worse for smaller-scale models. This suggests the emergence of CoT abilities for visual reasoning in LLMs at larger scales even when world knowledge is limited. Overall, we find limitations in the abilities of VLMs and LLMs for more complex visual reasoning, and highlight the important role that LLMs can play in visual reasoning.
LLMs are not Zero-Shot Reasoners for Biomedical Information Extraction
Aug 22, 2024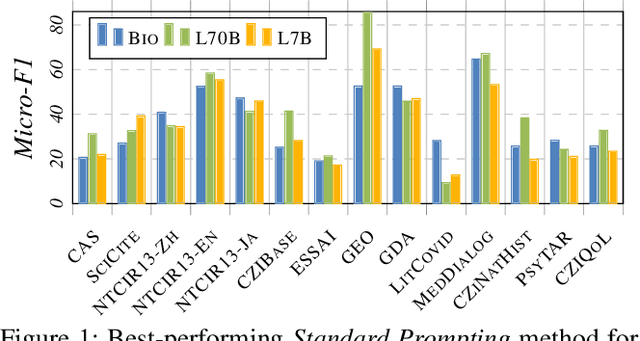
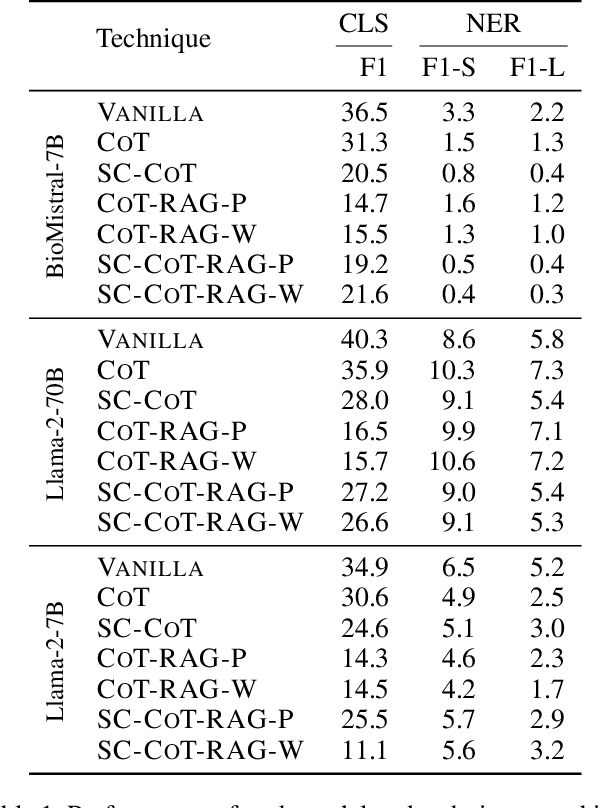
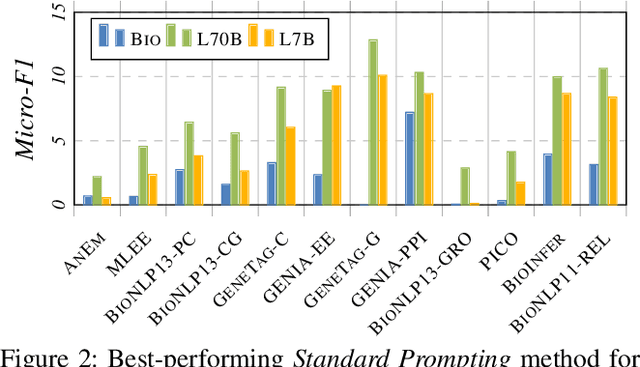
Abstract:Large Language Models (LLMs) are increasingly adopted for applications in healthcare, reaching the performance of domain experts on tasks such as question answering and document summarisation. Despite their success on these tasks, it is unclear how well LLMs perform on tasks that are traditionally pursued in the biomedical domain, such as structured information extration. To breach this gap, in this paper, we systematically benchmark LLM performance in Medical Classification and Named Entity Recognition (NER) tasks. We aim to disentangle the contribution of different factors to the performance, particularly the impact of LLMs' task knowledge and reasoning capabilities, their (parametric) domain knowledge, and addition of external knowledge. To this end we evaluate various open LLMs -- including BioMistral and Llama-2 models -- on a diverse set of biomedical datasets, using standard prompting, Chain-of-Thought (CoT) and Self-Consistency based reasoning as well as Retrieval-Augmented Generation (RAG) with PubMed and Wikipedia corpora. Counter-intuitively, our results reveal that standard prompting consistently outperforms more complex techniques across both tasks, laying bare the limitations in the current application of CoT, self-consistency and RAG in the biomedical domain. Our findings suggest that advanced prompting methods developed for knowledge- or reasoning-intensive tasks, such as CoT or RAG, are not easily portable to biomedical tasks where precise structured outputs are required. This highlights the need for more effective integration of external knowledge and reasoning mechanisms in LLMs to enhance their performance in real-world biomedical applications.
uMedSum: A Unified Framework for Advancing Medical Abstractive Summarization
Aug 22, 2024



Abstract:Medical abstractive summarization faces the challenge of balancing faithfulness and informativeness. Current methods often sacrifice key information for faithfulness or introduce confabulations when prioritizing informativeness. While recent advancements in techniques like in-context learning (ICL) and fine-tuning have improved medical summarization, they often overlook crucial aspects such as faithfulness and informativeness without considering advanced methods like model reasoning and self-improvement. Moreover, the field lacks a unified benchmark, hindering systematic evaluation due to varied metrics and datasets. This paper addresses these gaps by presenting a comprehensive benchmark of six advanced abstractive summarization methods across three diverse datasets using five standardized metrics. Building on these findings, we propose uMedSum, a modular hybrid summarization framework that introduces novel approaches for sequential confabulation removal followed by key missing information addition, ensuring both faithfulness and informativeness. Our work improves upon previous GPT-4-based state-of-the-art (SOTA) medical summarization methods, significantly outperforming them in both quantitative metrics and qualitative domain expert evaluations. Notably, we achieve an average relative performance improvement of 11.8% in reference-free metrics over the previous SOTA. Doctors prefer uMedSum's summaries 6 times more than previous SOTA in difficult cases where there are chances of confabulations or missing information. These results highlight uMedSum's effectiveness and generalizability across various datasets and metrics, marking a significant advancement in medical summarization.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge