Viktor Schlegel
SynBench: A Benchmark for Differentially Private Text Generation
Sep 18, 2025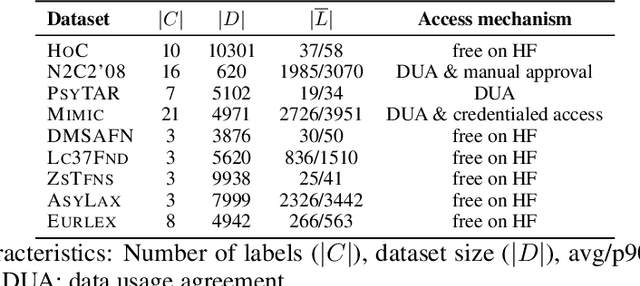
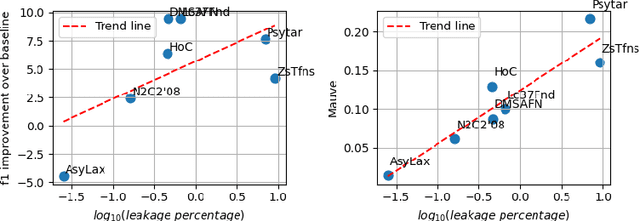
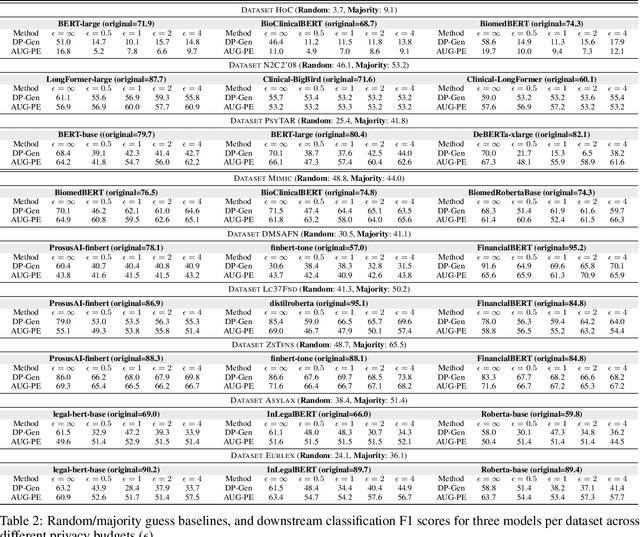
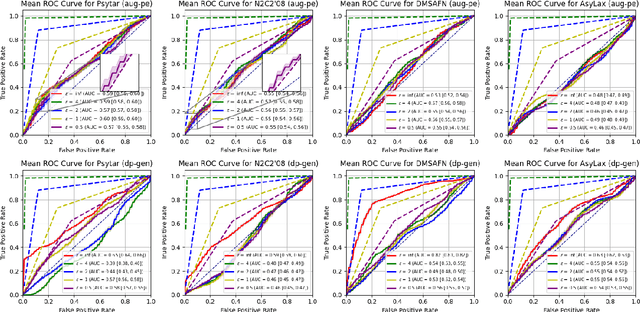
Abstract:Data-driven decision support in high-stakes domains like healthcare and finance faces significant barriers to data sharing due to regulatory, institutional, and privacy concerns. While recent generative AI models, such as large language models, have shown impressive performance in open-domain tasks, their adoption in sensitive environments remains limited by unpredictable behaviors and insufficient privacy-preserving datasets for benchmarking. Existing anonymization methods are often inadequate, especially for unstructured text, as redaction and masking can still allow re-identification. Differential Privacy (DP) offers a principled alternative, enabling the generation of synthetic data with formal privacy assurances. In this work, we address these challenges through three key contributions. First, we introduce a comprehensive evaluation framework with standardized utility and fidelity metrics, encompassing nine curated datasets that capture domain-specific complexities such as technical jargon, long-context dependencies, and specialized document structures. Second, we conduct a large-scale empirical study benchmarking state-of-the-art DP text generation methods and LLMs of varying sizes and different fine-tuning strategies, revealing that high-quality domain-specific synthetic data generation under DP constraints remains an unsolved challenge, with performance degrading as domain complexity increases. Third, we develop a membership inference attack (MIA) methodology tailored for synthetic text, providing first empirical evidence that the use of public datasets - potentially present in pre-training corpora - can invalidate claimed privacy guarantees. Our findings underscore the urgent need for rigorous privacy auditing and highlight persistent gaps between open-domain and specialist evaluations, informing responsible deployment of generative AI in privacy-sensitive, high-stakes settings.
MIRA: Medical Time Series Foundation Model for Real-World Health Data
Jun 09, 2025
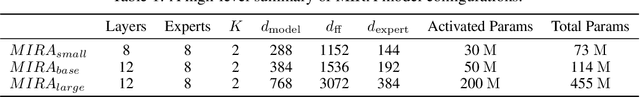

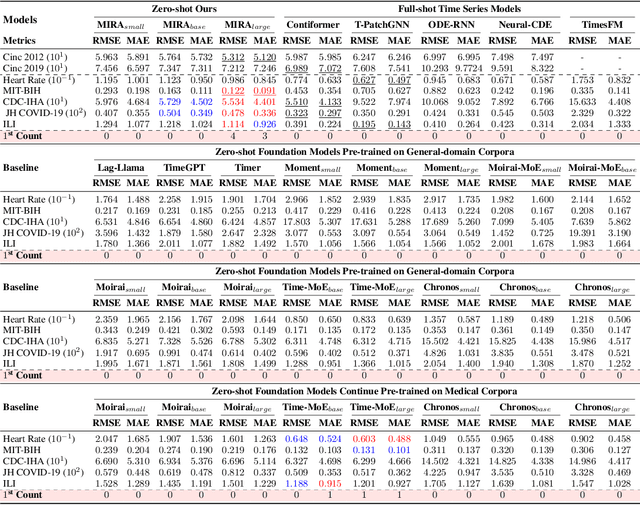
Abstract:A unified foundation model for medical time series -- pretrained on open access and ethics board-approved medical corpora -- offers the potential to reduce annotation burdens, minimize model customization, and enable robust transfer across clinical institutions, modalities, and tasks, particularly in data-scarce or privacy-constrained environments. However, existing generalist time series foundation models struggle to handle medical time series data due to their inherent challenges, including irregular intervals, heterogeneous sampling rates, and frequent missing values. To address these challenges, we introduce MIRA, a unified foundation model specifically designed for medical time series forecasting. MIRA incorporates a Continuous-Time Rotary Positional Encoding that enables fine-grained modeling of variable time intervals, a frequency-specific mixture-of-experts layer that routes computation across latent frequency regimes to further promote temporal specialization, and a Continuous Dynamics Extrapolation Block based on Neural ODE that models the continuous trajectory of latent states, enabling accurate forecasting at arbitrary target timestamps. Pretrained on a large-scale and diverse medical corpus comprising over 454 billion time points collect from publicly available datasets, MIRA achieves reductions in forecasting errors by an average of 10% and 7% in out-of-distribution and in-distribution scenarios, respectively, when compared to other zero-shot and fine-tuned baselines. We also introduce a comprehensive benchmark spanning multiple downstream clinical tasks, establishing a foundation for future research in medical time series modeling.
Generating Synthetic Data with Formal Privacy Guarantees: State of the Art and the Road Ahead
Mar 26, 2025Abstract:Privacy-preserving synthetic data offers a promising solution to harness segregated data in high-stakes domains where information is compartmentalized for regulatory, privacy, or institutional reasons. This survey provides a comprehensive framework for understanding the landscape of privacy-preserving synthetic data, presenting the theoretical foundations of generative models and differential privacy followed by a review of state-of-the-art methods across tabular data, images, and text. Our synthesis of evaluation approaches highlights the fundamental trade-off between utility for down-stream tasks and privacy guarantees, while identifying critical research gaps: the lack of realistic benchmarks representing specialized domains and insufficient empirical evaluations required to contextualise formal guarantees. Through empirical analysis of four leading methods on five real-world datasets from specialized domains, we demonstrate significant performance degradation under realistic privacy constraints ($\epsilon \leq 4$), revealing a substantial gap between results reported on general domain benchmarks and performance on domain-specific data. %Our findings highlight key challenges including unaccounted privacy leakage, insufficient empirical verification of formal guarantees, and a critical deficit of realistic benchmarks. These challenges underscore the need for robust evaluation frameworks, standardized benchmarks for specialized domains, and improved techniques to address the unique requirements of privacy-sensitive fields such that this technology can deliver on its considerable potential.
BRIDGE: Bootstrapping Text to Control Time-Series Generation via Multi-Agent Iterative Optimization and Diffusion Modelling
Mar 05, 2025



Abstract:Time-series Generation (TSG) is a prominent research area with broad applications in simulations, data augmentation, and counterfactual analysis. While existing methods have shown promise in unconditional single-domain TSG, real-world applications demand for cross-domain approaches capable of controlled generation tailored to domain-specific constraints and instance-level requirements. In this paper, we argue that text can provide semantic insights, domain information and instance-specific temporal patterns, to guide and improve TSG. We introduce ``Text-Controlled TSG'', a task focused on generating realistic time series by incorporating textual descriptions. To address data scarcity in this setting, we propose a novel LLM-based Multi-Agent framework that synthesizes diverse, realistic text-to-TS datasets. Furthermore, we introduce BRIDGE, a hybrid text-controlled TSG framework that integrates semantic prototypes with text description for supporting domain-level guidance. This approach achieves state-of-the-art generation fidelity on 11 of 12 datasets, and improves controllability by 12.52% on MSE and 6.34% MAE compared to no text input generation, highlighting its potential for generating tailored time-series data.
Pay Attention to Real World Perturbations! Natural Robustness Evaluation in Machine Reading Comprehension
Feb 23, 2025Abstract:As neural language models achieve human-comparable performance on Machine Reading Comprehension (MRC) and see widespread adoption, ensuring their robustness in real-world scenarios has become increasingly important. Current robustness evaluation research, though, primarily develops synthetic perturbation methods, leaving unclear how well they reflect real life scenarios. Considering this, we present a framework to automatically examine MRC models on naturally occurring textual perturbations, by replacing paragraph in MRC benchmarks with their counterparts based on available Wikipedia edit history. Such perturbation type is natural as its design does not stem from an arteficial generative process, inherently distinct from the previously investigated synthetic approaches. In a large-scale study encompassing SQUAD datasets and various model architectures we observe that natural perturbations result in performance degradation in pre-trained encoder language models. More worryingly, these state-of-the-art Flan-T5 and Large Language Models (LLMs) inherit these errors. Further experiments demonstrate that our findings generalise to natural perturbations found in other more challenging MRC benchmarks. In an effort to mitigate these errors, we show that it is possible to improve the robustness to natural perturbations by training on naturally or synthetically perturbed examples, though a noticeable gap still remains compared to performance on unperturbed data.
Learning to Generate and Evaluate Fact-checking Explanations with Transformers
Oct 21, 2024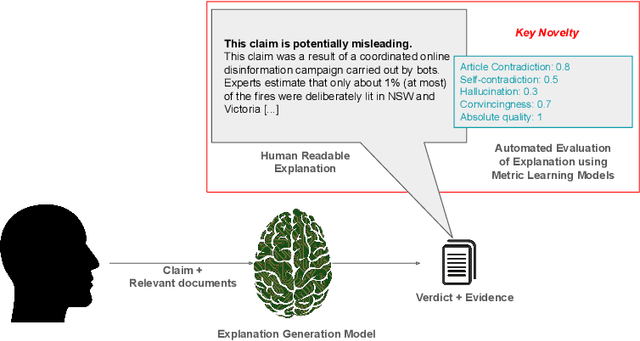

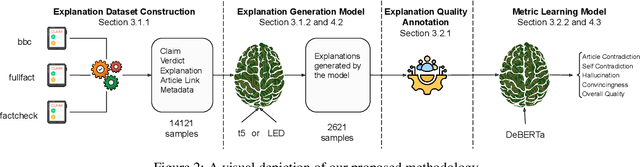
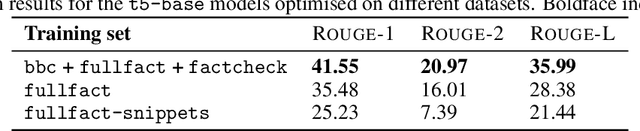
Abstract:In an era increasingly dominated by digital platforms, the spread of misinformation poses a significant challenge, highlighting the need for solutions capable of assessing information veracity. Our research contributes to the field of Explainable Artificial Antelligence (XAI) by developing transformer-based fact-checking models that contextualise and justify their decisions by generating human-accessible explanations. Importantly, we also develop models for automatic evaluation of explanations for fact-checking verdicts across different dimensions such as \texttt{(self)-contradiction}, \texttt{hallucination}, \texttt{convincingness} and \texttt{overall quality}. By introducing human-centred evaluation methods and developing specialised datasets, we emphasise the need for aligning Artificial Intelligence (AI)-generated explanations with human judgements. This approach not only advances theoretical knowledge in XAI but also holds practical implications by enhancing the transparency, reliability and users' trust in AI-driven fact-checking systems. Furthermore, the development of our metric learning models is a first step towards potentially increasing efficiency and reducing reliance on extensive manual assessment. Based on experimental results, our best performing generative model \textsc{ROUGE-1} score of 47.77, demonstrating superior performance in generating fact-checking explanations, particularly when provided with high-quality evidence. Additionally, the best performing metric learning model showed a moderately strong correlation with human judgements on objective dimensions such as \texttt{(self)-contradiction and \texttt{hallucination}, achieving a Matthews Correlation Coefficient (MCC) of around 0.7.}
Representation Learning of Structured Data for Medical Foundation Models
Oct 17, 2024

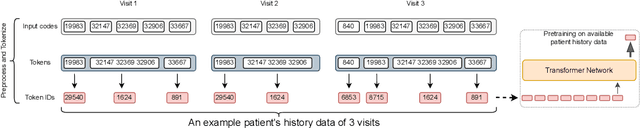
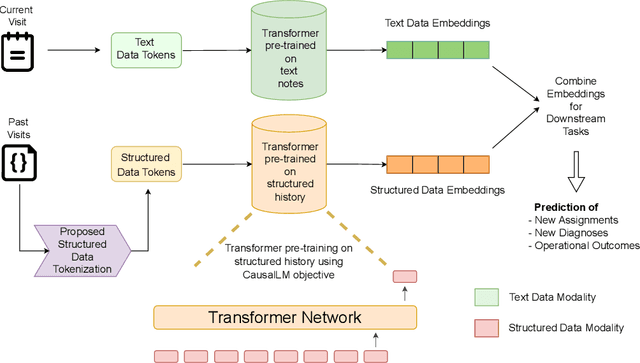
Abstract:Large Language Models (LLMs) have demonstrated remarkable performance across various domains, including healthcare. However, their ability to effectively represent structured non-textual data, such as the alphanumeric medical codes used in records like ICD-10 or SNOMED-CT, is limited and has been particularly exposed in recent research. This paper examines the challenges LLMs face in processing medical codes due to the shortcomings of current tokenization methods. As a result, we introduce the UniStruct architecture to design a multimodal medical foundation model of unstructured text and structured data, which addresses these challenges by adapting subword tokenization techniques specifically for the structured medical codes. Our approach is validated through model pre-training on both an extensive internal medical database and a public repository of structured medical records. Trained on over 1 billion tokens on the internal medical database, the proposed model achieves up to a 23% improvement in evaluation metrics, with around 2% gain attributed to our proposed tokenization. Additionally, when evaluated on the EHRSHOT public benchmark with a 1/1000 fraction of the pre-training data, the UniStruct model improves performance on over 42% of the downstream tasks. Our approach not only enhances the representation and generalization capabilities of patient-centric models but also bridges a critical gap in representation learning models' ability to handle complex structured medical data, alongside unstructured text.
Seemingly Plausible Distractors in Multi-Hop Reasoning: Are Large Language Models Attentive Readers?
Sep 08, 2024Abstract:State-of-the-art Large Language Models (LLMs) are accredited with an increasing number of different capabilities, ranging from reading comprehension, over advanced mathematical and reasoning skills to possessing scientific knowledge. In this paper we focus on their multi-hop reasoning capability: the ability to identify and integrate information from multiple textual sources. Given the concerns with the presence of simplifying cues in existing multi-hop reasoning benchmarks, which allow models to circumvent the reasoning requirement, we set out to investigate, whether LLMs are prone to exploiting such simplifying cues. We find evidence that they indeed circumvent the requirement to perform multi-hop reasoning, but they do so in more subtle ways than what was reported about their fine-tuned pre-trained language model (PLM) predecessors. Motivated by this finding, we propose a challenging multi-hop reasoning benchmark, by generating seemingly plausible multi-hop reasoning chains, which ultimately lead to incorrect answers. We evaluate multiple open and proprietary state-of-the-art LLMs, and find that their performance to perform multi-hop reasoning is affected, as indicated by up to 45% relative decrease in F1 score when presented with such seemingly plausible alternatives. We conduct a deeper analysis and find evidence that while LLMs tend to ignore misleading lexical cues, misleading reasoning paths indeed present a significant challenge.
MEDSAGE: Enhancing Robustness of Medical Dialogue Summarization to ASR Errors with LLM-generated Synthetic Dialogues
Aug 26, 2024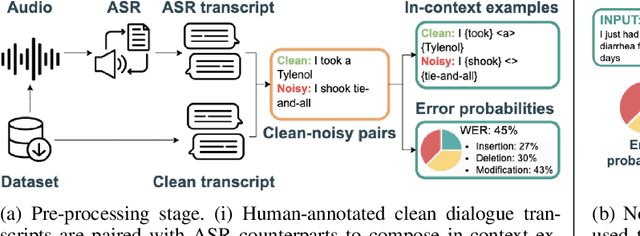
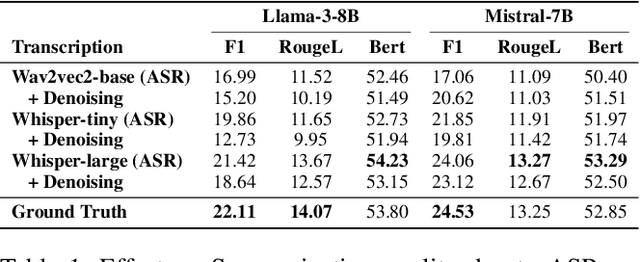

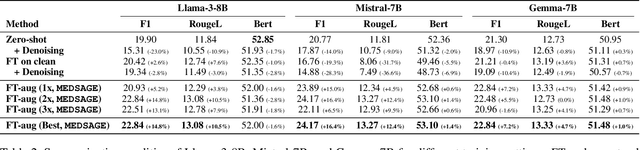
Abstract:Automatic Speech Recognition (ASR) systems are pivotal in transcribing speech into text, yet the errors they introduce can significantly degrade the performance of downstream tasks like summarization. This issue is particularly pronounced in clinical dialogue summarization, a low-resource domain where supervised data for fine-tuning is scarce, necessitating the use of ASR models as black-box solutions. Employing conventional data augmentation for enhancing the noise robustness of summarization models is not feasible either due to the unavailability of sufficient medical dialogue audio recordings and corresponding ASR transcripts. To address this challenge, we propose MEDSAGE, an approach for generating synthetic samples for data augmentation using Large Language Models (LLMs). Specifically, we leverage the in-context learning capabilities of LLMs and instruct them to generate ASR-like errors based on a few available medical dialogue examples with audio recordings. Experimental results show that LLMs can effectively model ASR noise, and incorporating this noisy data into the training process significantly improves the robustness and accuracy of medical dialogue summarization systems. This approach addresses the challenges of noisy ASR outputs in critical applications, offering a robust solution to enhance the reliability of clinical dialogue summarization.
uMedSum: A Unified Framework for Advancing Medical Abstractive Summarization
Aug 22, 2024



Abstract:Medical abstractive summarization faces the challenge of balancing faithfulness and informativeness. Current methods often sacrifice key information for faithfulness or introduce confabulations when prioritizing informativeness. While recent advancements in techniques like in-context learning (ICL) and fine-tuning have improved medical summarization, they often overlook crucial aspects such as faithfulness and informativeness without considering advanced methods like model reasoning and self-improvement. Moreover, the field lacks a unified benchmark, hindering systematic evaluation due to varied metrics and datasets. This paper addresses these gaps by presenting a comprehensive benchmark of six advanced abstractive summarization methods across three diverse datasets using five standardized metrics. Building on these findings, we propose uMedSum, a modular hybrid summarization framework that introduces novel approaches for sequential confabulation removal followed by key missing information addition, ensuring both faithfulness and informativeness. Our work improves upon previous GPT-4-based state-of-the-art (SOTA) medical summarization methods, significantly outperforming them in both quantitative metrics and qualitative domain expert evaluations. Notably, we achieve an average relative performance improvement of 11.8% in reference-free metrics over the previous SOTA. Doctors prefer uMedSum's summaries 6 times more than previous SOTA in difficult cases where there are chances of confabulations or missing information. These results highlight uMedSum's effectiveness and generalizability across various datasets and metrics, marking a significant advancement in medical summarization.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge