Ali Parviz
M^4olGen: Multi-Agent, Multi-Stage Molecular Generation under Precise Multi-Property Constraints
Jan 16, 2026Abstract:Generating molecules that satisfy precise numeric constraints over multiple physicochemical properties is critical and challenging. Although large language models (LLMs) are expressive, they struggle with precise multi-objective control and numeric reasoning without external structure and feedback. We introduce \textbf{M olGen}, a fragment-level, retrieval-augmented, two-stage framework for molecule generation under multi-property constraints. Stage I : Prototype generation: a multi-agent reasoner performs retrieval-anchored, fragment-level edits to produce a candidate near the feasible region. Stage II : RL-based fine-grained optimization: a fragment-level optimizer trained with Group Relative Policy Optimization (GRPO) applies one- or multi-hop refinements to explicitly minimize the property errors toward our target while regulating edit complexity and deviation from the prototype. A large, automatically curated dataset with reasoning chains of fragment edits and measured property deltas underpins both stages, enabling deterministic, reproducible supervision and controllable multi-hop reasoning. Unlike prior work, our framework better reasons about molecules by leveraging fragments and supports controllable refinement toward numeric targets. Experiments on generation under two sets of property constraints (QED, LogP, Molecular Weight and HOMO, LUMO) show consistent gains in validity and precise satisfaction of multi-property targets, outperforming strong LLMs and graph-based algorithms.
LLMs Can't Play Hangman: On the Necessity of a Private Working Memory for Language Agents
Jan 11, 2026Abstract:As LLMs move from text completion toward autonomous agents, they remain constrained by the standard chat interface, which lacks private working memory. This raises a fundamental question: can agents reliably perform interactive tasks that depend on hidden state? We define Private State Interactive Tasks (PSITs), which require agents to generate and maintain hidden information while producing consistent public responses. We show theoretically that any agent restricted to the public conversation history cannot simultaneously preserve secrecy and consistency in PSITs, yielding an impossibility theorem. To empirically validate this limitation, we introduce a self-consistency testing protocol that evaluates whether agents can maintain a hidden secret across forked dialogue branches. Standard chat-based LLMs and retrieval-based memory baselines fail this test regardless of scale, demonstrating that semantic retrieval does not enable true state maintenance. To address this, we propose a novel architecture incorporating an explicit private working memory; we demonstrate that this mechanism restores consistency, establishing private state as a necessary component for interactive language agents.
Best of Both Worlds: Advantages of Hybrid Graph Sequence Models
Nov 23, 2024Abstract:Modern sequence models (e.g., Transformers, linear RNNs, etc.) emerged as dominant backbones of recent deep learning frameworks, mainly due to their efficiency, representational power, and/or ability to capture long-range dependencies. Adopting these sequence models for graph-structured data has recently gained popularity as the alternative to Message Passing Neural Networks (MPNNs). There is, however, a lack of a common foundation about what constitutes a good graph sequence model, and a mathematical description of the benefits and deficiencies in adopting different sequence models for learning on graphs. To this end, we first present Graph Sequence Model (GSM), a unifying framework for adopting sequence models for graphs, consisting of three main steps: (1) Tokenization, which translates the graph into a set of sequences; (2) Local Encoding, which encodes local neighborhoods around each node; and (3) Global Encoding, which employs a scalable sequence model to capture long-range dependencies within the sequences. This framework allows us to understand, evaluate, and compare the power of different sequence model backbones in graph tasks. Our theoretical evaluations of the representation power of Transformers and modern recurrent models through the lens of global and local graph tasks show that there are both negative and positive sides for both types of models. Building on this observation, we present GSM++, a fast hybrid model that uses the Hierarchical Affinity Clustering (HAC) algorithm to tokenize the graph into hierarchical sequences, and then employs a hybrid architecture of Transformer to encode these sequences. Our theoretical and experimental results support the design of GSM++, showing that GSM++ outperforms baselines in most benchmark evaluations.
TGB 2.0: A Benchmark for Learning on Temporal Knowledge Graphs and Heterogeneous Graphs
Jun 14, 2024Abstract:Multi-relational temporal graphs are powerful tools for modeling real-world data, capturing the evolving and interconnected nature of entities over time. Recently, many novel models are proposed for ML on such graphs intensifying the need for robust evaluation and standardized benchmark datasets. However, the availability of such resources remains scarce and evaluation faces added complexity due to reproducibility issues in experimental protocols. To address these challenges, we introduce Temporal Graph Benchmark 2.0 (TGB 2.0), a novel benchmarking framework tailored for evaluating methods for predicting future links on Temporal Knowledge Graphs and Temporal Heterogeneous Graphs with a focus on large-scale datasets, extending the Temporal Graph Benchmark. TGB 2.0 facilitates comprehensive evaluations by presenting eight novel datasets spanning five domains with up to 53 million edges. TGB 2.0 datasets are significantly larger than existing datasets in terms of number of nodes, edges, or timestamps. In addition, TGB 2.0 provides a reproducible and realistic evaluation pipeline for multi-relational temporal graphs. Through extensive experimentation, we observe that 1) leveraging edge-type information is crucial to obtain high performance, 2) simple heuristic baselines are often competitive with more complex methods, 3) most methods fail to run on our largest datasets, highlighting the need for research on more scalable methods.
$\texttt{MiniMol}$: A Parameter-Efficient Foundation Model for Molecular Learning
Apr 23, 2024Abstract:In biological tasks, data is rarely plentiful as it is generated from hard-to-gather measurements. Therefore, pre-training foundation models on large quantities of available data and then transfer to low-data downstream tasks is a promising direction. However, how to design effective foundation models for molecular learning remains an open question, with existing approaches typically focusing on models with large parameter capacities. In this work, we propose $\texttt{MiniMol}$, a foundational model for molecular learning with 10 million parameters. $\texttt{MiniMol}$ is pre-trained on a mix of roughly 3300 sparsely defined graph- and node-level tasks of both quantum and biological nature. The pre-training dataset includes approximately 6 million molecules and 500 million labels. To demonstrate the generalizability of $\texttt{MiniMol}$ across tasks, we evaluate it on downstream tasks from the Therapeutic Data Commons (TDC) ADMET group showing significant improvements over the prior state-of-the-art foundation model across 17 tasks. $\texttt{MiniMol}$ will be a public and open-sourced model for future research.
Reducing the Cost of Quantum Chemical Data By Backpropagating Through Density Functional Theory
Feb 06, 2024Abstract:Density Functional Theory (DFT) accurately predicts the quantum chemical properties of molecules, but scales as $O(N_{\text{electrons}}^3)$. Sch\"utt et al. (2019) successfully approximate DFT 1000x faster with Neural Networks (NN). Arguably, the biggest problem one faces when scaling to larger molecules is the cost of DFT labels. For example, it took years to create the PCQ dataset (Nakata & Shimazaki, 2017) on which subsequent NNs are trained within a week. DFT labels molecules by minimizing energy $E(\cdot )$ as a "loss function." We bypass dataset creation by directly training NNs with $E(\cdot )$ as a loss function. For comparison, Sch\"utt et al. (2019) spent 626 hours creating a dataset on which they trained their NN for 160h, for a total of 786h; our method achieves comparable performance within 31h.
Resource-constrained knowledge diffusion processes inspired by human peer learning
Dec 01, 2023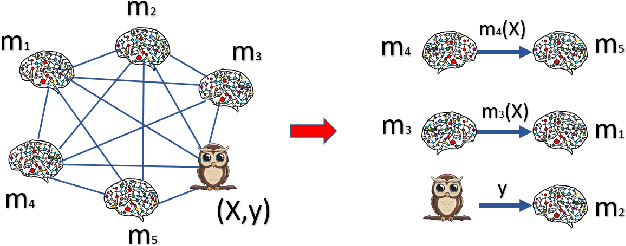
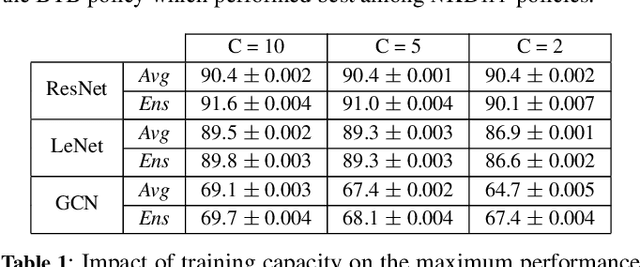
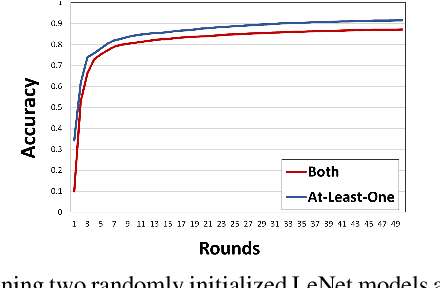
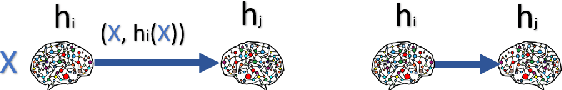
Abstract:We consider a setting where a population of artificial learners is given, and the objective is to optimize aggregate measures of performance, under constraints on training resources. The problem is motivated by the study of peer learning in human educational systems. In this context, we study natural knowledge diffusion processes in networks of interacting artificial learners. By `natural', we mean processes that reflect human peer learning where the students' internal state and learning process is mostly opaque, and the main degree of freedom lies in the formation of peer learning groups by a coordinator who can potentially evaluate the learners before assigning them to peer groups. Among else, we empirically show that such processes indeed make effective use of the training resources, and enable the design of modular neural models that have the capacity to generalize without being prone to overfitting noisy labels.
Towards Foundational Models for Molecular Learning on Large-Scale Multi-Task Datasets
Oct 18, 2023



Abstract:Recently, pre-trained foundation models have enabled significant advancements in multiple fields. In molecular machine learning, however, where datasets are often hand-curated, and hence typically small, the lack of datasets with labeled features, and codebases to manage those datasets, has hindered the development of foundation models. In this work, we present seven novel datasets categorized by size into three distinct categories: ToyMix, LargeMix and UltraLarge. These datasets push the boundaries in both the scale and the diversity of supervised labels for molecular learning. They cover nearly 100 million molecules and over 3000 sparsely defined tasks, totaling more than 13 billion individual labels of both quantum and biological nature. In comparison, our datasets contain 300 times more data points than the widely used OGB-LSC PCQM4Mv2 dataset, and 13 times more than the quantum-only QM1B dataset. In addition, to support the development of foundational models based on our proposed datasets, we present the Graphium graph machine learning library which simplifies the process of building and training molecular machine learning models for multi-task and multi-level molecular datasets. Finally, we present a range of baseline results as a starting point of multi-task and multi-level training on these datasets. Empirically, we observe that performance on low-resource biological datasets show improvement by also training on large amounts of quantum data. This indicates that there may be potential in multi-task and multi-level training of a foundation model and fine-tuning it to resource-constrained downstream tasks.
Generalized Laplacian Positional Encoding for Graph Representation Learning
Nov 10, 2022



Abstract:Graph neural networks (GNNs) are the primary tool for processing graph-structured data. Unfortunately, the most commonly used GNNs, called Message Passing Neural Networks (MPNNs) suffer from several fundamental limitations. To overcome these limitations, recent works have adapted the idea of positional encodings to graph data. This paper draws inspiration from the recent success of Laplacian-based positional encoding and defines a novel family of positional encoding schemes for graphs. We accomplish this by generalizing the optimization problem that defines the Laplace embedding to more general dissimilarity functions rather than the 2-norm used in the original formulation. This family of positional encodings is then instantiated by considering p-norms. We discuss a method for calculating these positional encoding schemes, implement it in PyTorch and demonstrate how the resulting positional encoding captures different properties of the graph. Furthermore, we demonstrate that this novel family of positional encodings can improve the expressive power of MPNNs. Lastly, we present preliminary experimental results.
Long Range Graph Benchmark
Jun 16, 2022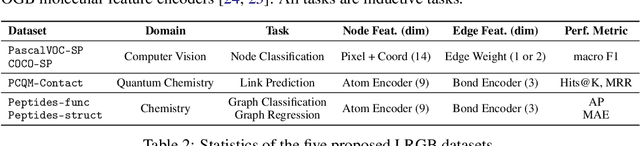
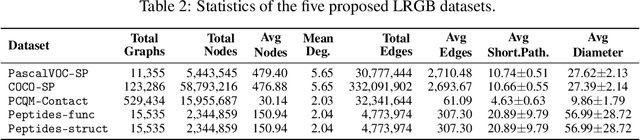
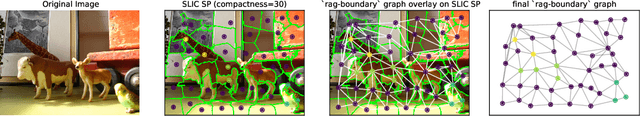
Abstract:Graph Neural Networks (GNNs) that are based on the message passing (MP) paradigm exchange information between 1-hop neighbors to build node representations at each layer. In principle, such networks are not able to capture long-range interactions (LRI) that may be desired or necessary for learning a given task on graphs. Recently, there has been an increasing interest in development of Transformer-based methods for graphs that can consider full node connectivity beyond the original sparse structure, thus enabling the modeling of LRI. However, MP-GNNs that simply rely on 1-hop message passing often fare better in several existing graph benchmarks when combined with positional feature representations, among other innovations, hence limiting the perceived utility and ranking of Transformer-like architectures. Here, we present the Long Range Graph Benchmark (LRGB) with 5 graph learning datasets: PascalVOC-SP, COCO-SP, PCQM-Contact, Peptides-func and Peptides-struct that arguably require LRI reasoning to achieve strong performance in a given task. We benchmark both baseline GNNs and Graph Transformer networks to verify that the models which capture long-range dependencies perform significantly better on these tasks. Therefore, these datasets are suitable for benchmarking and exploration of MP-GNNs and Graph Transformer architectures that are intended to capture LRI.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge