Andrew Fitzgibbon
On Stochastic Rounding with Few Random Bits
Apr 29, 2025Abstract:Large-scale numerical computations make increasing use of low-precision (LP) floating point formats and mixed precision arithmetic, which can be enhanced by the technique of stochastic rounding (SR), that is, rounding an intermediate high-precision value up or down randomly as a function of the value's distance to the two rounding candidates. Stochastic rounding requires, in addition to the high-precision input value, a source of random bits. As the provision of high-quality random bits is an additional computational cost, it is of interest to require as few bits as possible while maintaining the desirable properties of SR in a given computation, or computational domain. This paper examines a number of possible implementations of few-bit stochastic rounding (FBSR), and shows how several natural implementations can introduce sometimes significant bias into the rounding process, which are not present in the case of infinite-bit, infinite-precision examinations of these implementations. The paper explores the impact of these biases in machine learning examples, and hence opens another class of configuration parameters of which practitioners should be aware when developing or adopting low-precision floating point. Code is available at http://github.com/graphcore-research/arith25-stochastic-rounding.
Scalify: scale propagation for efficient low-precision LLM training
Jul 24, 2024



Abstract:Low-precision formats such as float8 have been introduced in machine learning accelerated hardware to improve computational efficiency for large language models training and inference. Nevertheless, adoption by the ML community has been slowed down by the complex, and sometimes brittle, techniques required to match higher precision training accuracy. In this work, we present Scalify, a end-to-end scale propagation paradigm for computational graphs, generalizing and formalizing existing tensor scaling methods. Experiment results show that Scalify supports out-of-the-box float8 matrix multiplication and gradients representation, as well as float16 optimizer state storage. Our JAX implementation of Scalify is open-sourced at https://github.com/graphcore-research/jax-scalify
MESS: Modern Electronic Structure Simulations
Jun 05, 2024Abstract:Electronic structure simulation (ESS) has been used for decades to provide quantitative scientific insights on an atomistic scale, enabling advances in chemistry, biology, and materials science, among other disciplines. Following standard practice in scientific computing, the software packages driving these studies have been implemented in compiled languages such as FORTRAN and C. However, the recent introduction of machine learning (ML) into these domains has meant that ML models must be coded in these languages, or that complex software bridges have to be built between ML models in Python and these large compiled software systems. This is in contrast with recent progress in modern ML frameworks which aim to optimise both ease of use and high performance by harnessing hardware acceleration of tensor programs defined in Python. We introduce MESS: a modern electronic structure simulation package implemented in JAX; porting the ESS code to the ML world. We outline the costs and benefits of following the software development practices used in ML for this important scientific workload. MESS shows significant speedups n widely available hardware accelerators and simultaneously opens a clear pathway towards combining ESS with ML. MESS is available at https://github.com/graphcore-research/mess.
$\texttt{MiniMol}$: A Parameter-Efficient Foundation Model for Molecular Learning
Apr 23, 2024Abstract:In biological tasks, data is rarely plentiful as it is generated from hard-to-gather measurements. Therefore, pre-training foundation models on large quantities of available data and then transfer to low-data downstream tasks is a promising direction. However, how to design effective foundation models for molecular learning remains an open question, with existing approaches typically focusing on models with large parameter capacities. In this work, we propose $\texttt{MiniMol}$, a foundational model for molecular learning with 10 million parameters. $\texttt{MiniMol}$ is pre-trained on a mix of roughly 3300 sparsely defined graph- and node-level tasks of both quantum and biological nature. The pre-training dataset includes approximately 6 million molecules and 500 million labels. To demonstrate the generalizability of $\texttt{MiniMol}$ across tasks, we evaluate it on downstream tasks from the Therapeutic Data Commons (TDC) ADMET group showing significant improvements over the prior state-of-the-art foundation model across 17 tasks. $\texttt{MiniMol}$ will be a public and open-sourced model for future research.
Generating QM1B with PySCF$_{\text{IPU}}$
Nov 02, 2023Abstract:The emergence of foundation models in Computer Vision and Natural Language Processing have resulted in immense progress on downstream tasks. This progress was enabled by datasets with billions of training examples. Similar benefits are yet to be unlocked for quantum chemistry, where the potential of deep learning is constrained by comparatively small datasets with 100k to 20M training examples. These datasets are limited in size because the labels are computed using the accurate (but computationally demanding) predictions of Density Functional Theory (DFT). Notably, prior DFT datasets were created using CPU supercomputers without leveraging hardware acceleration. In this paper, we take a first step towards utilising hardware accelerators by introducing the data generator PySCF$_{\text{IPU}}$ using Intelligence Processing Units (IPUs). This allowed us to create the dataset QM1B with one billion training examples containing 9-11 heavy atoms. We demonstrate that a simple baseline neural network (SchNet 9M) improves its performance by simply increasing the amount of training data without additional inductive biases. To encourage future researchers to use QM1B responsibly, we highlight several limitations of QM1B and emphasise the low-resolution of our DFT options, which also serves as motivation for even larger, more accurate datasets. Code and dataset are available on Github: http://github.com/graphcore-research/pyscf-ipu
Towards Foundational Models for Molecular Learning on Large-Scale Multi-Task Datasets
Oct 18, 2023



Abstract:Recently, pre-trained foundation models have enabled significant advancements in multiple fields. In molecular machine learning, however, where datasets are often hand-curated, and hence typically small, the lack of datasets with labeled features, and codebases to manage those datasets, has hindered the development of foundation models. In this work, we present seven novel datasets categorized by size into three distinct categories: ToyMix, LargeMix and UltraLarge. These datasets push the boundaries in both the scale and the diversity of supervised labels for molecular learning. They cover nearly 100 million molecules and over 3000 sparsely defined tasks, totaling more than 13 billion individual labels of both quantum and biological nature. In comparison, our datasets contain 300 times more data points than the widely used OGB-LSC PCQM4Mv2 dataset, and 13 times more than the quantum-only QM1B dataset. In addition, to support the development of foundational models based on our proposed datasets, we present the Graphium graph machine learning library which simplifies the process of building and training molecular machine learning models for multi-task and multi-level molecular datasets. Finally, we present a range of baseline results as a starting point of multi-task and multi-level training on these datasets. Empirically, we observe that performance on low-resource biological datasets show improvement by also training on large amounts of quantum data. This indicates that there may be potential in multi-task and multi-level training of a foundation model and fine-tuning it to resource-constrained downstream tasks.
GPS++: Reviving the Art of Message Passing for Molecular Property Prediction
Feb 06, 2023



Abstract:We present GPS++, a hybrid Message Passing Neural Network / Graph Transformer model for molecular property prediction. Our model integrates a well-tuned local message passing component and biased global attention with other key ideas from prior literature to achieve state-of-the-art results on large-scale molecular dataset PCQM4Mv2. Through a thorough ablation study we highlight the impact of individual components and, contrary to expectations set by recent trends, find that nearly all of the model's performance can be maintained without any use of global self-attention. We also show that our approach is significantly more accurate than prior art when 3D positional information is not available.
Efficient and Sound Differentiable Programming in a Functional Array-Processing Language
Dec 20, 2022Abstract:Automatic differentiation (AD) is a technique for computing the derivative of a function represented by a program. This technique is considered as the de-facto standard for computing the differentiation in many machine learning and optimisation software tools. Despite the practicality of this technique, the performance of the differentiated programs, especially for functional languages and in the presence of vectors, is suboptimal. We present an AD system for a higher-order functional array-processing language. The core functional language underlying this system simultaneously supports both source-to-source forward-mode AD and global optimisations such as loop transformations. In combination, gradient computation with forward-mode AD can be as efficient as reverse mode, and the Jacobian matrices required for numerical algorithms such as Gauss-Newton and Levenberg-Marquardt can be efficiently computed.
BESS: Balanced Entity Sampling and Sharing for Large-Scale Knowledge Graph Completion
Nov 22, 2022
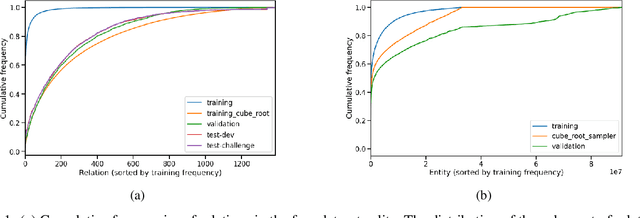
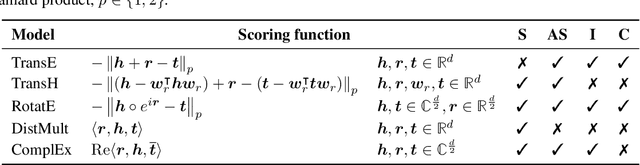
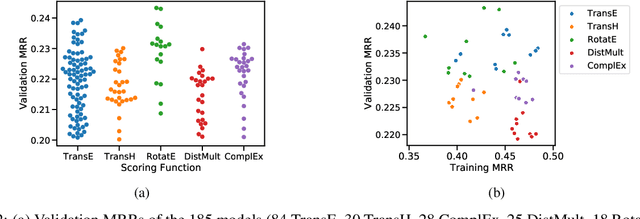
Abstract:We present the award-winning submission to the WikiKG90Mv2 track of OGB-LSC@NeurIPS 2022. The task is link-prediction on the large-scale knowledge graph WikiKG90Mv2, consisting of 90M+ nodes and 600M+ edges. Our solution uses a diverse ensemble of $85$ Knowledge Graph Embedding models combining five different scoring functions (TransE, TransH, RotatE, DistMult, ComplEx) and two different loss functions (log-sigmoid, sampled softmax cross-entropy). Each individual model is trained in parallel on a Graphcore Bow Pod$_{16}$ using BESS (Balanced Entity Sampling and Sharing), a new distribution framework for KGE training and inference based on balanced collective communications between workers. Our final model achieves a validation MRR of 0.2922 and a test-challenge MRR of 0.2562, winning the first place in the competition. The code is publicly available at: https://github.com/graphcore/distributed-kge-poplar/tree/2022-ogb-submission.
FLAG: Flow-based 3D Avatar Generation from Sparse Observations
Mar 11, 2022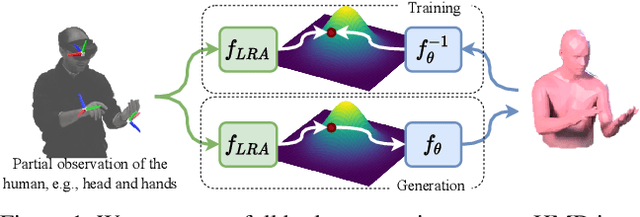
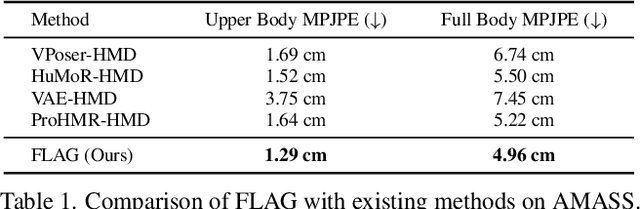
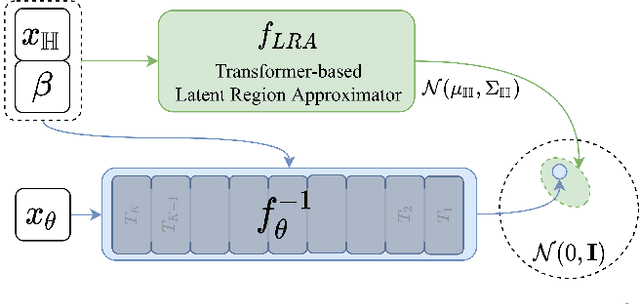
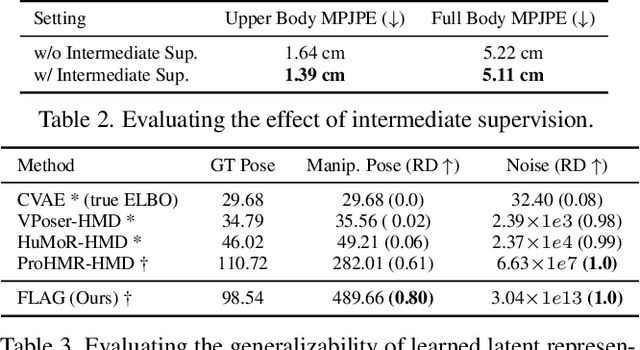
Abstract:To represent people in mixed reality applications for collaboration and communication, we need to generate realistic and faithful avatar poses. However, the signal streams that can be applied for this task from head-mounted devices (HMDs) are typically limited to head pose and hand pose estimates. While these signals are valuable, they are an incomplete representation of the human body, making it challenging to generate a faithful full-body avatar. We address this challenge by developing a flow-based generative model of the 3D human body from sparse observations, wherein we learn not only a conditional distribution of 3D human pose, but also a probabilistic mapping from observations to the latent space from which we can generate a plausible pose along with uncertainty estimates for the joints. We show that our approach is not only a strong predictive model, but can also act as an efficient pose prior in different optimization settings where a good initial latent code plays a major role.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge