Kerstin Kläser
$\texttt{MiniMol}$: A Parameter-Efficient Foundation Model for Molecular Learning
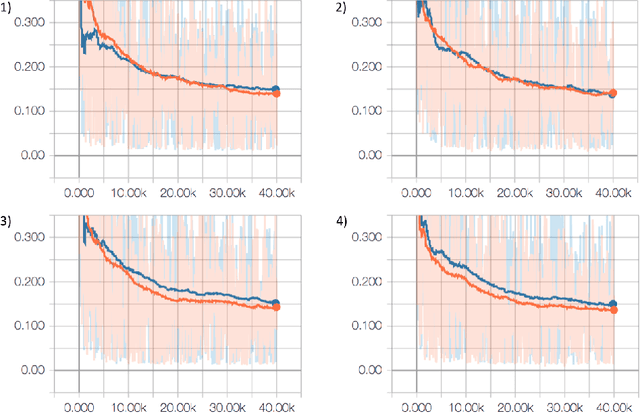
Apr 23, 2024Abstract:In biological tasks, data is rarely plentiful as it is generated from hard-to-gather measurements. Therefore, pre-training foundation models on large quantities of available data and then transfer to low-data downstream tasks is a promising direction. However, how to design effective foundation models for molecular learning remains an open question, with existing approaches typically focusing on models with large parameter capacities. In this work, we propose $\texttt{MiniMol}$, a foundational model for molecular learning with 10 million parameters. $\texttt{MiniMol}$ is pre-trained on a mix of roughly 3300 sparsely defined graph- and node-level tasks of both quantum and biological nature. The pre-training dataset includes approximately 6 million molecules and 500 million labels. To demonstrate the generalizability of $\texttt{MiniMol}$ across tasks, we evaluate it on downstream tasks from the Therapeutic Data Commons (TDC) ADMET group showing significant improvements over the prior state-of-the-art foundation model across 17 tasks. $\texttt{MiniMol}$ will be a public and open-sourced model for future research.
Combining multimodal information for Metal Artefact Reduction: An unsupervised deep learning framework
Apr 20, 2020
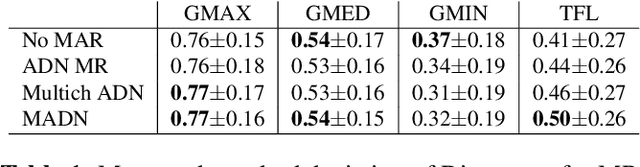

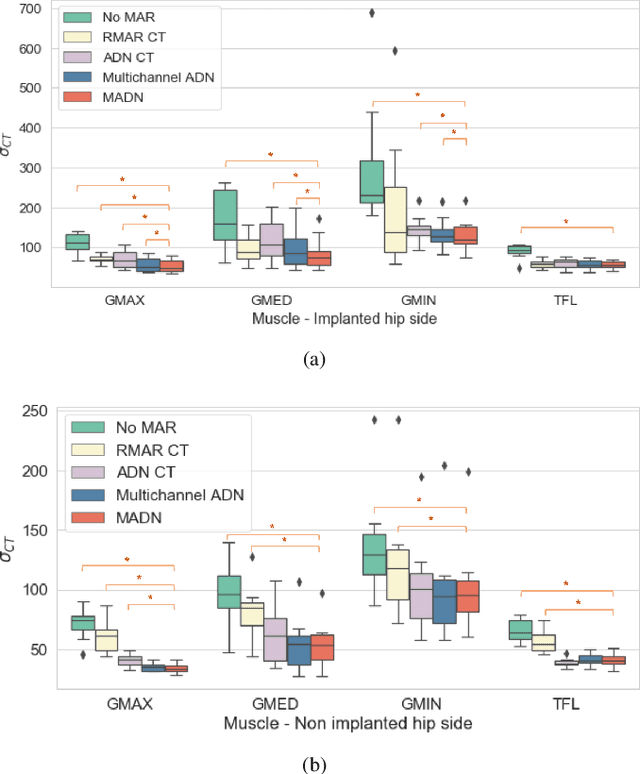
Abstract:Metal artefact reduction (MAR) techniques aim at removing metal-induced noise from clinical images. In Computed Tomography (CT), supervised deep learning approaches have been shown effective but limited in generalisability, as they mostly rely on synthetic data. In Magnetic Resonance Imaging (MRI) instead, no method has yet been introduced to correct the susceptibility artefact, still present even in MAR-specific acquisitions. In this work, we hypothesise that a multimodal approach to MAR would improve both CT and MRI. Given their different artefact appearance, their complementary information can compensate for the corrupted signal in either modality. We thus propose an unsupervised deep learning method for multimodal MAR. We introduce the use of Locally Normalised Cross Correlation as a loss term to encourage the fusion of multimodal information. Experiments show that our approach favours a smoother correction in the CT, while promoting signal recovery in the MRI.
Improved MR to CT synthesis for PET/MR attenuation correction using Imitation Learning
Aug 27, 2019Abstract:The ability to synthesise Computed Tomography images - commonly known as pseudo CT, or pCT - from MRI input data is commonly assessed using an intensity-wise similarity, such as an L2-norm between the ground truth CT and the pCT. However, given that the ultimate purpose is often to use the pCT as an attenuation map ($\mu$-map) in Positron Emission Tomography Magnetic Resonance Imaging (PET/MRI), minimising the error between pCT and CT is not necessarily optimal. The main objective should be to predict a pCT that, when used as $\mu$-map, reconstructs a pseudo PET (pPET) which is as close as possible to the gold standard PET. To this end, we propose a novel multi-hypothesis deep learning framework that generates pCTs by minimising a combination of the pixel-wise error between pCT and CT and a proposed metric-loss that itself is represented by a convolutional neural network (CNN) and aims to minimise subsequent PET residuals. The model is trained on a database of 400 paired MR/CT/PET image slices. Quantitative results show that the network generates pCTs that seem less accurate when evaluating the Mean Absolute Error on the pCT (69.68HU) compared to a baseline CNN (66.25HU), but lead to significant improvement in the PET reconstruction - 115a.u. compared to baseline 140a.u.
Deep Boosted Regression for MR to CT Synthesis
Aug 22, 2018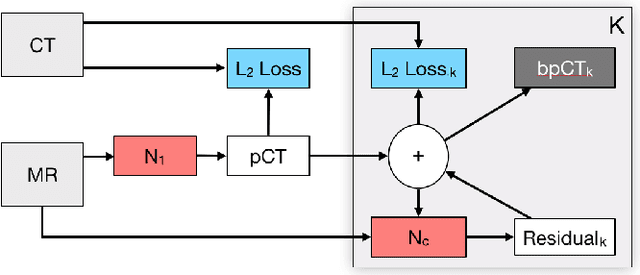
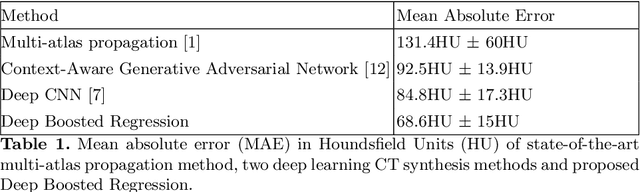
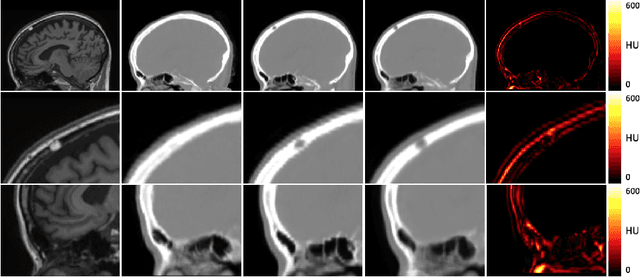
Abstract:Attenuation correction is an essential requirement of positron emission tomography (PET) image reconstruction to allow for accurate quantification. However, attenuation correction is particularly challenging for PET-MRI as neither PET nor magnetic resonance imaging (MRI) can directly image tissue attenuation properties. MRI-based computed tomography (CT) synthesis has been proposed as an alternative to physics based and segmentation-based approaches that assign a population-based tissue density value in order to generate an attenuation map. We propose a novel deep fully convolutional neural network that generates synthetic CTs in a recursive manner by gradually reducing the residuals of the previous network, increasing the overall accuracy and generalisability, while keeping the number of trainable parameters within reasonable limits. The model is trained on a database of 20 pre-acquired MRI/CT pairs and a four-fold random bootstrapped validation with a 80:20 split is performed. Quantitative results show that the proposed framework outperforms a state-of-the-art atlas-based approach decreasing the Mean Absolute Error (MAE) from 131HU to 68HU for the synthetic CTs and reducing the PET reconstruction error from 14.3% to 7.2%.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge