Luis Müller
Generalizable Insights for Graph Transformers in Theory and Practice
Nov 11, 2025Abstract:Graph Transformers (GTs) have shown strong empirical performance, yet current architectures vary widely in their use of attention mechanisms, positional embeddings (PEs), and expressivity. Existing expressivity results are often tied to specific design choices and lack comprehensive empirical validation on large-scale data. This leaves a gap between theory and practice, preventing generalizable insights that exceed particular application domains. Here, we propose the Generalized-Distance Transformer (GDT), a GT architecture using standard attention that incorporates many advancements for GTs from recent years, and develop a fine-grained understanding of the GDT's representation power in terms of attention and PEs. Through extensive experiments, we identify design choices that consistently perform well across various applications, tasks, and model scales, demonstrating strong performance in a few-shot transfer setting without fine-tuning. Our evaluation covers over eight million graphs with roughly 270M tokens across diverse domains, including image-based object detection, molecular property prediction, code summarization, and out-of-distribution algorithmic reasoning. We distill our theoretical and practical findings into several generalizable insights about effective GT design, training, and inference.
Position: Graph Learning Will Lose Relevance Due To Poor Benchmarks
Feb 20, 2025
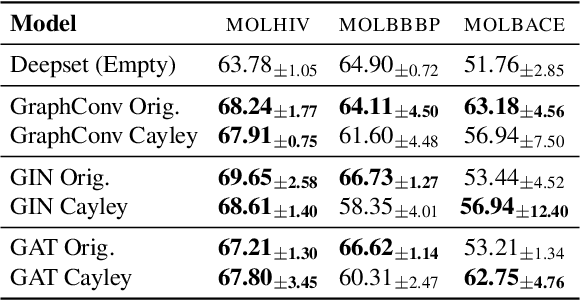
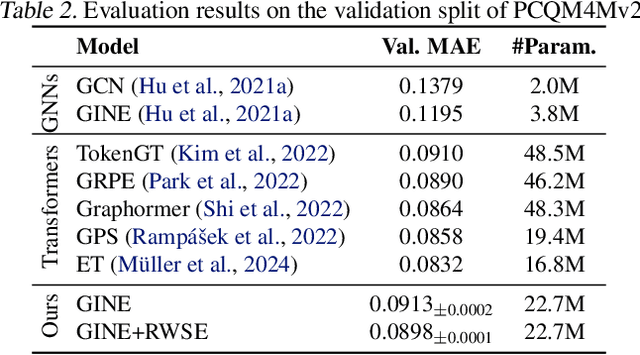
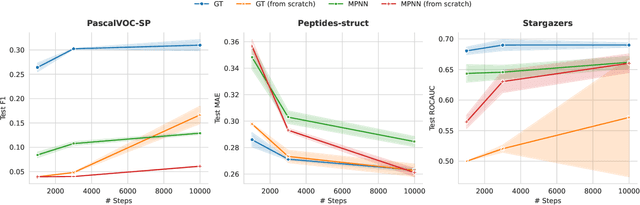
Abstract:While machine learning on graphs has demonstrated promise in drug design and molecular property prediction, significant benchmarking challenges hinder its further progress and relevance. Current benchmarking practices often lack focus on transformative, real-world applications, favoring narrow domains like two-dimensional molecular graphs over broader, impactful areas such as combinatorial optimization, relational databases, or chip design. Additionally, many benchmark datasets poorly represent the underlying data, leading to inadequate abstractions and misaligned use cases. Fragmented evaluations and an excessive focus on accuracy further exacerbate these issues, incentivizing overfitting rather than fostering generalizable insights. These limitations have prevented the development of truly useful graph foundation models. This position paper calls for a paradigm shift toward more meaningful benchmarks, rigorous evaluation protocols, and stronger collaboration with domain experts to drive impactful and reliable advances in graph learning research, unlocking the potential of graph learning.
Aligning Transformers with Weisfeiler-Leman
Jun 05, 2024



Abstract:Graph neural network architectures aligned with the $k$-dimensional Weisfeiler--Leman ($k$-WL) hierarchy offer theoretically well-understood expressive power. However, these architectures often fail to deliver state-of-the-art predictive performance on real-world graphs, limiting their practical utility. While recent works aligning graph transformer architectures with the $k$-WL hierarchy have shown promising empirical results, employing transformers for higher orders of $k$ remains challenging due to a prohibitive runtime and memory complexity of self-attention as well as impractical architectural assumptions, such as an infeasible number of attention heads. Here, we advance the alignment of transformers with the $k$-WL hierarchy, showing stronger expressivity results for each $k$, making them more feasible in practice. In addition, we develop a theoretical framework that allows the study of established positional encodings such as Laplacian PEs and SPE. We evaluate our transformers on the large-scale PCQM4Mv2 dataset, showing competitive predictive performance with the state-of-the-art and demonstrating strong downstream performance when fine-tuning them on small-scale molecular datasets. Our code is available at https://github.com/luis-mueller/wl-transformers.
$\texttt{MiniMol}$: A Parameter-Efficient Foundation Model for Molecular Learning
Apr 23, 2024Abstract:In biological tasks, data is rarely plentiful as it is generated from hard-to-gather measurements. Therefore, pre-training foundation models on large quantities of available data and then transfer to low-data downstream tasks is a promising direction. However, how to design effective foundation models for molecular learning remains an open question, with existing approaches typically focusing on models with large parameter capacities. In this work, we propose $\texttt{MiniMol}$, a foundational model for molecular learning with 10 million parameters. $\texttt{MiniMol}$ is pre-trained on a mix of roughly 3300 sparsely defined graph- and node-level tasks of both quantum and biological nature. The pre-training dataset includes approximately 6 million molecules and 500 million labels. To demonstrate the generalizability of $\texttt{MiniMol}$ across tasks, we evaluate it on downstream tasks from the Therapeutic Data Commons (TDC) ADMET group showing significant improvements over the prior state-of-the-art foundation model across 17 tasks. $\texttt{MiniMol}$ will be a public and open-sourced model for future research.
Towards Principled Graph Transformers
Jan 18, 2024



Abstract:Graph learning architectures based on the k-dimensional Weisfeiler-Leman (k-WL) hierarchy offer a theoretically well-understood expressive power. However, such architectures often fail to deliver solid predictive performance on real-world tasks, limiting their practical impact. In contrast, global attention-based models such as graph transformers demonstrate strong performance in practice, but comparing their expressive power with the k-WL hierarchy remains challenging, particularly since these architectures rely on positional or structural encodings for their expressivity and predictive performance. To address this, we show that the recently proposed Edge Transformer, a global attention model operating on node pairs instead of nodes, has at least 3-WL expressive power. Empirically, we demonstrate that the Edge Transformer surpasses other theoretically aligned architectures regarding predictive performance while not relying on positional or structural encodings.
Towards Foundational Models for Molecular Learning on Large-Scale Multi-Task Datasets
Oct 18, 2023



Abstract:Recently, pre-trained foundation models have enabled significant advancements in multiple fields. In molecular machine learning, however, where datasets are often hand-curated, and hence typically small, the lack of datasets with labeled features, and codebases to manage those datasets, has hindered the development of foundation models. In this work, we present seven novel datasets categorized by size into three distinct categories: ToyMix, LargeMix and UltraLarge. These datasets push the boundaries in both the scale and the diversity of supervised labels for molecular learning. They cover nearly 100 million molecules and over 3000 sparsely defined tasks, totaling more than 13 billion individual labels of both quantum and biological nature. In comparison, our datasets contain 300 times more data points than the widely used OGB-LSC PCQM4Mv2 dataset, and 13 times more than the quantum-only QM1B dataset. In addition, to support the development of foundational models based on our proposed datasets, we present the Graphium graph machine learning library which simplifies the process of building and training molecular machine learning models for multi-task and multi-level molecular datasets. Finally, we present a range of baseline results as a starting point of multi-task and multi-level training on these datasets. Empirically, we observe that performance on low-resource biological datasets show improvement by also training on large amounts of quantum data. This indicates that there may be potential in multi-task and multi-level training of a foundation model and fine-tuning it to resource-constrained downstream tasks.
Attending to Graph Transformers
Feb 08, 2023Abstract:Recently, transformer architectures for graphs emerged as an alternative to established techniques for machine learning with graphs, such as graph neural networks. So far, they have shown promising empirical results, e.g., on molecular prediction datasets, often attributed to their ability to circumvent graph neural networks' shortcomings, such as over-smoothing and over-squashing. Here, we derive a taxonomy of graph transformer architectures, bringing some order to this emerging field. We overview their theoretical properties, survey structural and positional encodings, and discuss extensions for important graph classes, e.g., 3D molecular graphs. Empirically, we probe how well graph transformers can recover various graph properties, how well they can deal with heterophilic graphs, and to what extent they prevent over-squashing. Further, we outline open challenges and research direction to stimulate future work. Our code is available at https://github.com/luis-mueller/probing-graph-transformers.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge