Zhiyi Li
Orb: A Fast, Scalable Neural Network Potential
Oct 29, 2024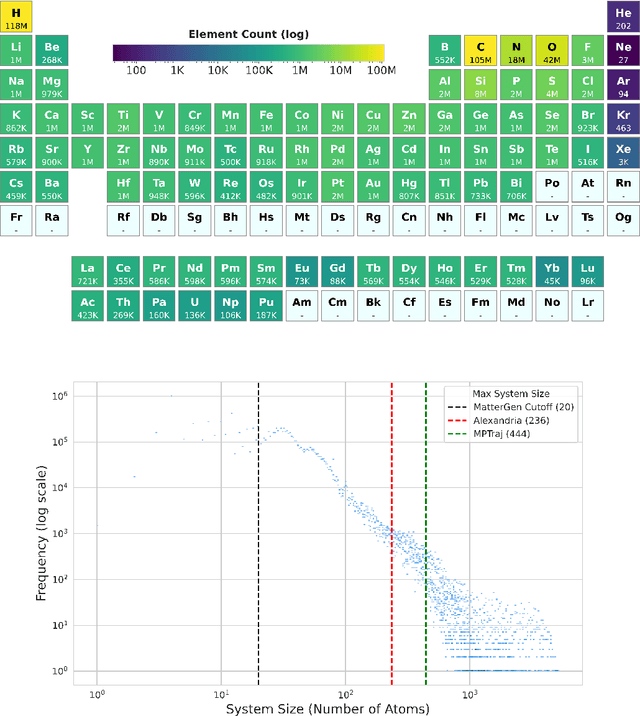
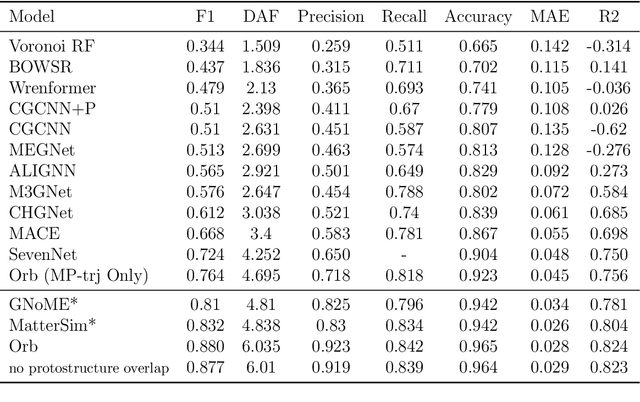
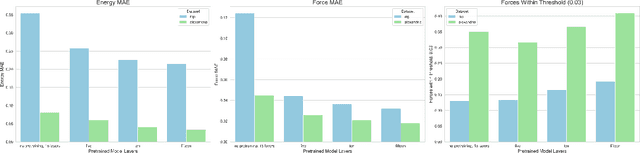
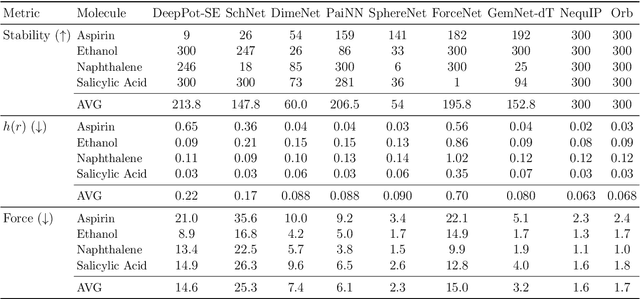
Abstract:We introduce Orb, a family of universal interatomic potentials for atomistic modelling of materials. Orb models are 3-6 times faster than existing universal potentials, stable under simulation for a range of out of distribution materials and, upon release, represented a 31% reduction in error over other methods on the Matbench Discovery benchmark. We explore several aspects of foundation model development for materials, with a focus on diffusion pretraining. We evaluate Orb as a model for geometry optimization, Monte Carlo and molecular dynamics simulations.
Comparative Analysis of Extrinsic Factors for NER in French
Oct 16, 2024



Abstract:Named entity recognition (NER) is a crucial task that aims to identify structured information, which is often replete with complex, technical terms and a high degree of variability. Accurate and reliable NER can facilitate the extraction and analysis of important information. However, NER for other than English is challenging due to limited data availability, as the high expertise, time, and expenses are required to annotate its data. In this paper, by using the limited data, we explore various factors including model structure, corpus annotation scheme and data augmentation techniques to improve the performance of a NER model for French. Our experiments demonstrate that these approaches can significantly improve the model's F1 score from original CRF score of 62.41 to 79.39. Our findings suggest that considering different extrinsic factors and combining these techniques is a promising approach for improving NER performance where the size of data is limited.
Statistical Properties of Robust Satisficing
May 30, 2024Abstract:The Robust Satisficing (RS) model is an emerging approach to robust optimization, offering streamlined procedures and robust generalization across various applications. However, the statistical theory of RS remains unexplored in the literature. This paper fills in the gap by comprehensively analyzing the theoretical properties of the RS model. Notably, the RS structure offers a more straightforward path to deriving statistical guarantees compared to the seminal Distributionally Robust Optimization (DRO), resulting in a richer set of results. In particular, we establish two-sided confidence intervals for the optimal loss without the need to solve a minimax optimization problem explicitly. We further provide finite-sample generalization error bounds for the RS optimizer. Importantly, our results extend to scenarios involving distribution shifts, where discrepancies exist between the sampling and target distributions. Our numerical experiments show that the RS model consistently outperforms the baseline empirical risk minimization in small-sample regimes and under distribution shifts. Furthermore, compared to the DRO model, the RS model exhibits lower sensitivity to hyperparameter tuning, highlighting its practicability for robustness considerations.
Obj-NeRF: Extract Object NeRFs from Multi-view Images
Nov 26, 2023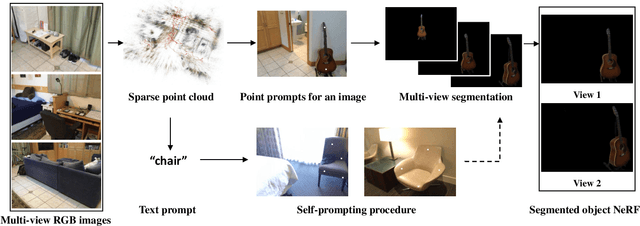
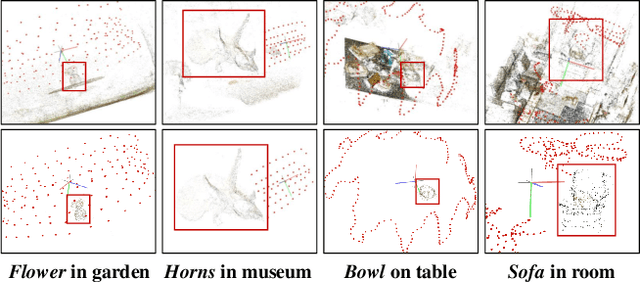
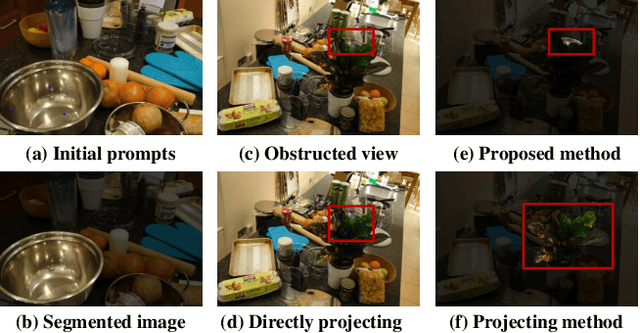
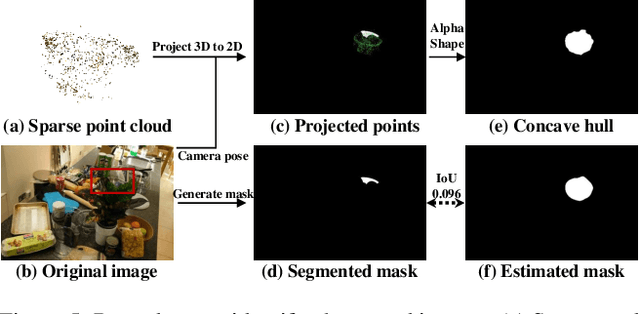
Abstract:Neural Radiance Fields (NeRFs) have demonstrated remarkable effectiveness in novel view synthesis within 3D environments. However, extracting a radiance field of one specific object from multi-view images encounters substantial challenges due to occlusion and background complexity, thereby presenting difficulties in downstream applications such as NeRF editing and 3D mesh extraction. To solve this problem, in this paper, we propose Obj-NeRF, a comprehensive pipeline that recovers the 3D geometry of a specific object from multi-view images using a single prompt. This method combines the 2D segmentation capabilities of the Segment Anything Model (SAM) in conjunction with the 3D reconstruction ability of NeRF. Specifically, we first obtain multi-view segmentation for the indicated object using SAM with a single prompt. Then, we use the segmentation images to supervise NeRF construction, integrating several effective techniques. Additionally, we construct a large object-level NeRF dataset containing diverse objects, which can be useful in various downstream tasks. To demonstrate the practicality of our method, we also apply Obj-NeRF to various applications, including object removal, rotation, replacement, and recoloring.
Enhancing Energy Efficiency for Reconfigurable Intelligent Surfaces with Practical Power Models
Oct 24, 2023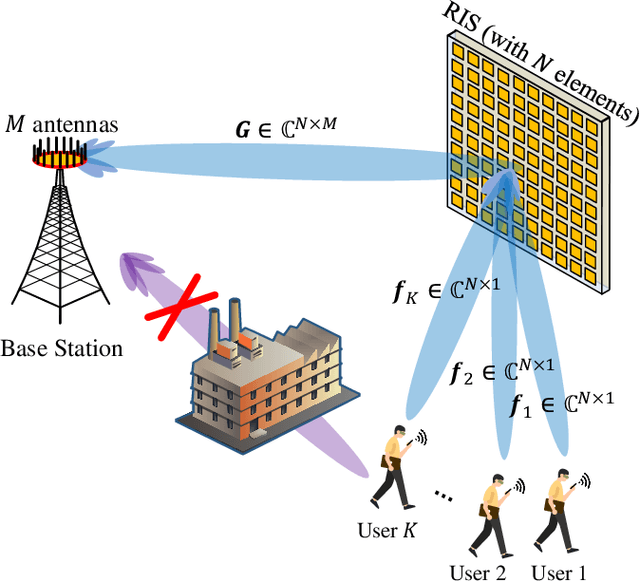
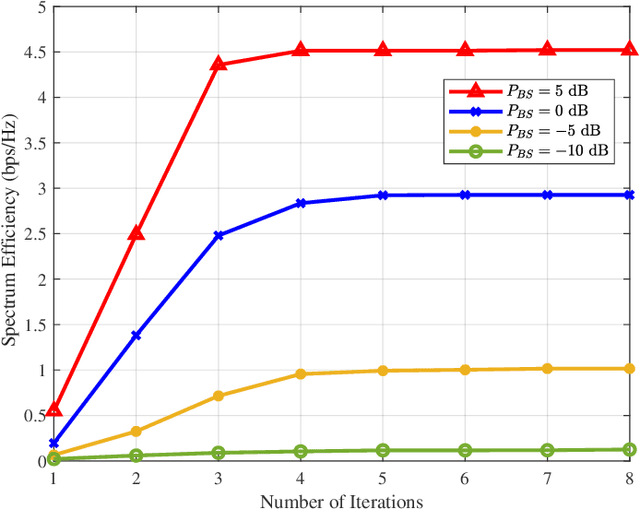
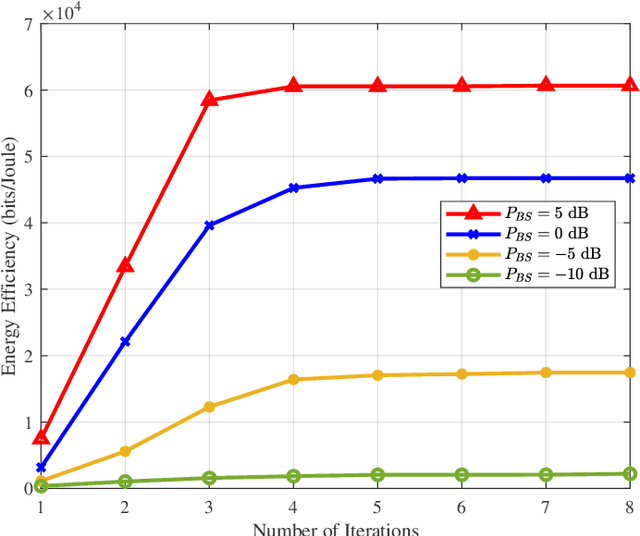
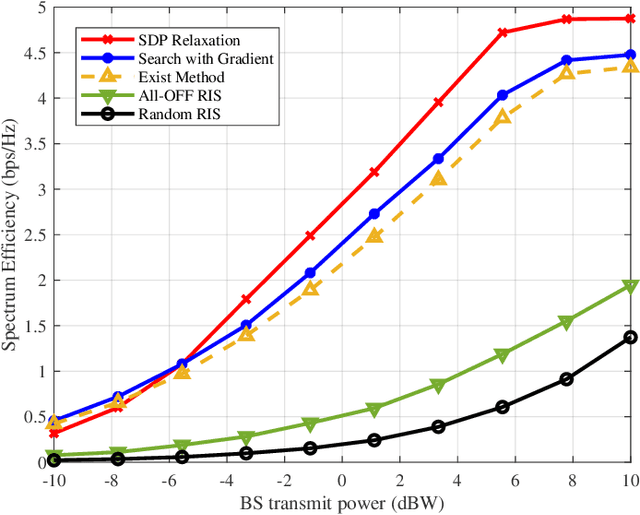
Abstract:Reconfigurable intelligent surfaces (RISs) are widely considered a promising technology for future wireless communication systems. As an important indicator of RIS-assisted communication systems in green wireless communications, energy efficiency (EE) has recently received intensive research interest as an optimization target. However, most previous works have ignored the different power consumption between ON and OFF states of the PIN diodes attached to each RIS element. This oversight results in extensive unnecessary power consumption and reduction of actual EE due to the inaccurate power model. To address this issue, in this paper, we first utilize a practical power model for a RIS-assisted multi-user multiple-input single-output (MU-MISO) communication system, which takes into account the difference in power dissipation caused by ON-OFF states of RIS's PIN diodes. Based on this model, we formulate a more accurate EE optimization problem. However, this problem is non-convex and has mixed-integer properties, which poses a challenge for optimization. To solve the problem, an effective alternating optimization (AO) algorithm framework is utilized to optimize the base station and RIS beamforming precoder separately. To obtain the essential RIS beamforming precoder, we develop two effective methods based on maximum gradient search and SDP relaxation respectively. Theoretical analysis shows the exponential complexity of the original problem has been reduced to polynomial complexity. Simulation results demonstrate that the proposed algorithm outperforms the existing ones, leading to a significant increase in EE across a diverse set of scenarios.
Towards Foundational Models for Molecular Learning on Large-Scale Multi-Task Datasets
Oct 18, 2023



Abstract:Recently, pre-trained foundation models have enabled significant advancements in multiple fields. In molecular machine learning, however, where datasets are often hand-curated, and hence typically small, the lack of datasets with labeled features, and codebases to manage those datasets, has hindered the development of foundation models. In this work, we present seven novel datasets categorized by size into three distinct categories: ToyMix, LargeMix and UltraLarge. These datasets push the boundaries in both the scale and the diversity of supervised labels for molecular learning. They cover nearly 100 million molecules and over 3000 sparsely defined tasks, totaling more than 13 billion individual labels of both quantum and biological nature. In comparison, our datasets contain 300 times more data points than the widely used OGB-LSC PCQM4Mv2 dataset, and 13 times more than the quantum-only QM1B dataset. In addition, to support the development of foundational models based on our proposed datasets, we present the Graphium graph machine learning library which simplifies the process of building and training molecular machine learning models for multi-task and multi-level molecular datasets. Finally, we present a range of baseline results as a starting point of multi-task and multi-level training on these datasets. Empirically, we observe that performance on low-resource biological datasets show improvement by also training on large amounts of quantum data. This indicates that there may be potential in multi-task and multi-level training of a foundation model and fine-tuning it to resource-constrained downstream tasks.
Extrinsic Factors Affecting the Accuracy of Biomedical NER
May 29, 2023Abstract:Biomedical named entity recognition (NER) is a critial task that aims to identify structured information in clinical text, which is often replete with complex, technical terms and a high degree of variability. Accurate and reliable NER can facilitate the extraction and analysis of important biomedical information, which can be used to improve downstream applications including the healthcare system. However, NER in the biomedical domain is challenging due to limited data availability, as the high expertise, time, and expenses are required to annotate its data. In this paper, by using the limited data, we explore various extrinsic factors including the corpus annotation scheme, data augmentation techniques, semi-supervised learning and Brill transformation, to improve the performance of a NER model on a clinical text dataset (i2b2 2012, \citet{sun-rumshisky-uzuner:2013}). Our experiments demonstrate that these approaches can significantly improve the model's F1 score from original 73.74 to 77.55. Our findings suggest that considering different extrinsic factors and combining these techniques is a promising approach for improving NER performance in the biomedical domain where the size of data is limited.
PopSparse: Accelerated block sparse matrix multiplication on IPU
Apr 05, 2023Abstract:Reducing the computational cost of running large scale neural networks using sparsity has attracted great attention in the deep learning community. While much success has been achieved in reducing FLOP and parameter counts while maintaining acceptable task performance, achieving actual speed improvements has typically been much more difficult, particularly on general purpose accelerators (GPAs) such as NVIDIA GPUs using low precision number formats. In this work we introduce PopSparse, a library that enables fast sparse operations on Graphcore IPUs by leveraging both the unique hardware characteristics of IPUs as well as any block structure defined in the data. We target two different types of sparsity: static, where the sparsity pattern is fixed at compile-time; and dynamic, where it can change each time the model is run. We present benchmark results for matrix multiplication for both of these modes on IPU with a range of block sizes, matrix sizes and densities. Results indicate that the PopSparse implementations are faster than dense matrix multiplications on IPU at a range of sparsity levels with large matrix size and block size. Furthermore, static sparsity in general outperforms dynamic sparsity. While previous work on GPAs has shown speedups only for very high sparsity (typically 99\% and above), the present work demonstrates that our static sparse implementation outperforms equivalent dense calculations in FP16 at lower sparsity (around 90%). IPU code is available to view and run at ipu.dev/sparsity-benchmarks, GPU code will be made available shortly.
GPS++: Reviving the Art of Message Passing for Molecular Property Prediction
Feb 06, 2023



Abstract:We present GPS++, a hybrid Message Passing Neural Network / Graph Transformer model for molecular property prediction. Our model integrates a well-tuned local message passing component and biased global attention with other key ideas from prior literature to achieve state-of-the-art results on large-scale molecular dataset PCQM4Mv2. Through a thorough ablation study we highlight the impact of individual components and, contrary to expectations set by recent trends, find that nearly all of the model's performance can be maintained without any use of global self-attention. We also show that our approach is significantly more accurate than prior art when 3D positional information is not available.
GPS++: An Optimised Hybrid MPNN/Transformer for Molecular Property Prediction
Dec 06, 2022



Abstract:This technical report presents GPS++, the first-place solution to the Open Graph Benchmark Large-Scale Challenge (OGB-LSC 2022) for the PCQM4Mv2 molecular property prediction task. Our approach implements several key principles from the prior literature. At its core our GPS++ method is a hybrid MPNN/Transformer model that incorporates 3D atom positions and an auxiliary denoising task. The effectiveness of GPS++ is demonstrated by achieving 0.0719 mean absolute error on the independent test-challenge PCQM4Mv2 split. Thanks to Graphcore IPU acceleration, GPS++ scales to deep architectures (16 layers), training at 3 minutes per epoch, and large ensemble (112 models), completing the final predictions in 1 hour 32 minutes, well under the 4 hour inference budget allocated. Our implementation is publicly available at: https://github.com/graphcore/ogb-lsc-pcqm4mv2.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge