Alberto Cattaneo
Ground-Truth Subgraphs for Better Training and Evaluation of Knowledge Graph Augmented LLMs
Nov 06, 2025Abstract:Retrieval of information from graph-structured knowledge bases represents a promising direction for improving the factuality of LLMs. While various solutions have been proposed, a comparison of methods is difficult due to the lack of challenging QA datasets with ground-truth targets for graph retrieval. We present SynthKGQA, a framework for generating high-quality synthetic Knowledge Graph Question Answering datasets from any Knowledge Graph, providing the full set of ground-truth facts in the KG to reason over each question. We show how, in addition to enabling more informative benchmarking of KG retrievers, the data produced with SynthKGQA also allows us to train better models. We apply SynthKGQA to Wikidata to generate GTSQA, a new dataset designed to test zero-shot generalization abilities of KG retrievers with respect to unseen graph structures and relation types, and benchmark popular solutions for KG-augmented LLMs on it.
Approximate Top-$k$ for Increased Parallelism
Dec 05, 2024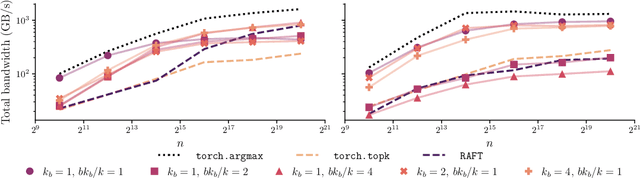
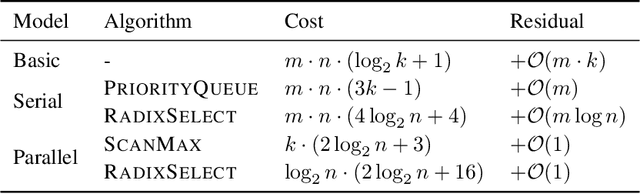
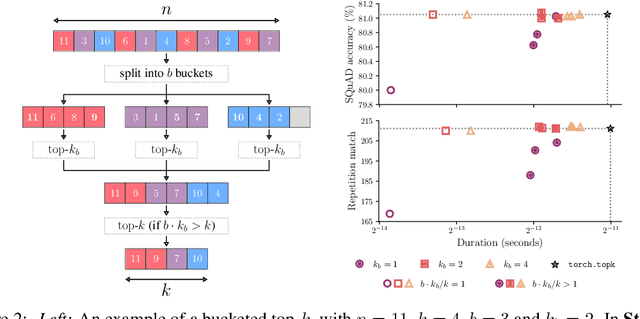
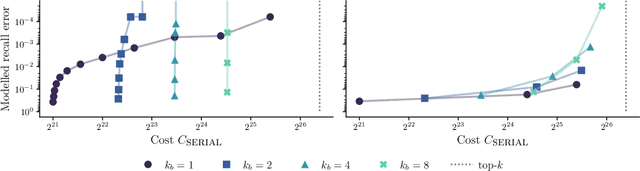
Abstract:We present an evaluation of bucketed approximate top-$k$ algorithms. Computing top-$k$ exactly suffers from limited parallelism, because the $k$ largest values must be aggregated along the vector, thus is not well suited to computation on highly-parallel machine learning accelerators. By relaxing the requirement that the top-$k$ is exact, bucketed algorithms can dramatically increase the parallelism available by independently computing many smaller top-$k$ operations. We explore the design choices of this class of algorithms using both theoretical analysis and empirical evaluation on downstream tasks. Our motivating examples are sparsity algorithms for language models, which often use top-$k$ to select the most important parameters or activations. We also release a fast bucketed top-$k$ implementation for PyTorch.
The Role of Graph Topology in the Performance of Biomedical Knowledge Graph Completion Models
Sep 06, 2024



Abstract:Knowledge Graph Completion has been increasingly adopted as a useful method for several tasks in biomedical research, like drug repurposing or drug-target identification. To that end, a variety of datasets and Knowledge Graph Embedding models has been proposed over the years. However, little is known about the properties that render a dataset useful for a given task and, even though theoretical properties of Knowledge Graph Embedding models are well understood, their practical utility in this field remains controversial. We conduct a comprehensive investigation into the topological properties of publicly available biomedical Knowledge Graphs and establish links to the accuracy observed in real-world applications. By releasing all model predictions and a new suite of analysis tools we invite the community to build upon our work and continue improving the understanding of these crucial applications.
BESS: Balanced Entity Sampling and Sharing for Large-Scale Knowledge Graph Completion
Nov 22, 2022
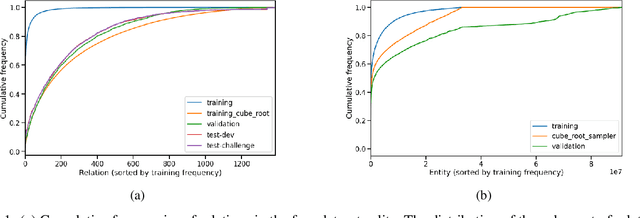
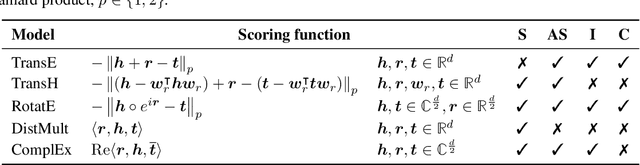
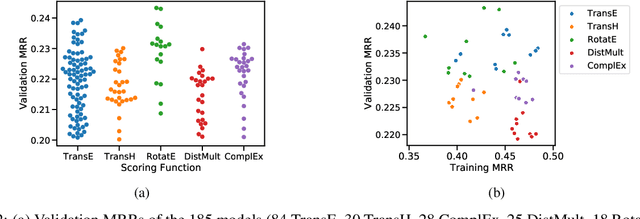
Abstract:We present the award-winning submission to the WikiKG90Mv2 track of OGB-LSC@NeurIPS 2022. The task is link-prediction on the large-scale knowledge graph WikiKG90Mv2, consisting of 90M+ nodes and 600M+ edges. Our solution uses a diverse ensemble of $85$ Knowledge Graph Embedding models combining five different scoring functions (TransE, TransH, RotatE, DistMult, ComplEx) and two different loss functions (log-sigmoid, sampled softmax cross-entropy). Each individual model is trained in parallel on a Graphcore Bow Pod$_{16}$ using BESS (Balanced Entity Sampling and Sharing), a new distribution framework for KGE training and inference based on balanced collective communications between workers. Our final model achieves a validation MRR of 0.2922 and a test-challenge MRR of 0.2562, winning the first place in the competition. The code is publicly available at: https://github.com/graphcore/distributed-kge-poplar/tree/2022-ogb-submission.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge