Junde Xu
From Supervision to Exploration: What Does Protein Language Model Learn During Reinforcement Learning?
Oct 02, 2025Abstract:Protein language models (PLMs) have advanced computational protein science through large-scale pretraining and scalable architectures. In parallel, reinforcement learning (RL) has broadened exploration and enabled precise multi-objective optimization in protein design. Yet whether RL can push PLMs beyond their pretraining priors to uncover latent sequence-structure-function rules remains unclear. We address this by pairing RL with PLMs across four domains: antimicrobial peptide design, kinase variant optimization, antibody engineering, and inverse folding. Using diverse RL algorithms and model classes, we ask if RL improves sampling efficiency and, more importantly, if it reveals capabilities not captured by supervised learning. Across benchmarks, RL consistently boosts success rates and sample efficiency. Performance follows a three-factor interaction: task headroom, reward fidelity, and policy capacity jointly determine gains. When rewards are accurate and informative, policies have sufficient capacity, and tasks leave room beyond supervised baselines, improvements scale; when rewards are noisy or capacity is constrained, gains saturate despite exploration. This view yields practical guidance for RL in protein design: prioritize reward modeling and calibration before scaling policy size, match algorithm and regularization strength to task difficulty, and allocate capacity where marginal gains are largest. Implementation is available at https://github.com/chq1155/RL-PLM.
DPPMask: Masked Image Modeling with Determinantal Point Processes
Mar 25, 2023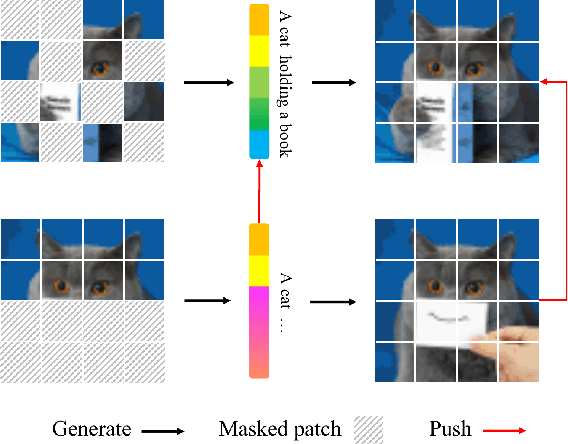
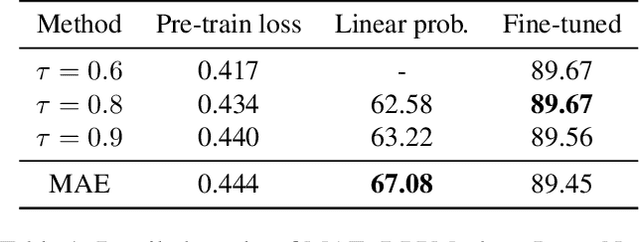
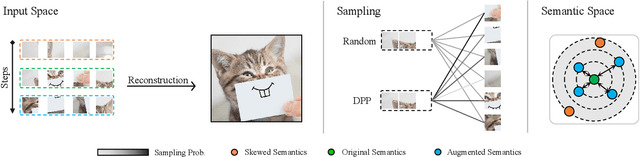
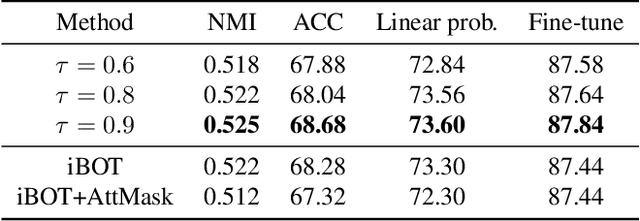
Abstract:Masked Image Modeling (MIM) has achieved impressive representative performance with the aim of reconstructing randomly masked images. Despite the empirical success, most previous works have neglected the important fact that it is unreasonable to force the model to reconstruct something beyond recovery, such as those masked objects. In this work, we show that uniformly random masking widely used in previous works unavoidably loses some key objects and changes original semantic information, resulting in a misalignment problem and hurting the representative learning eventually. To address this issue, we augment MIM with a new masking strategy namely the DPPMask by substituting the random process with Determinantal Point Process (DPPs) to reduce the semantic change of the image after masking. Our method is simple yet effective and requires no extra learnable parameters when implemented within various frameworks. In particular, we evaluate our method on two representative MIM frameworks, MAE and iBOT. We show that DPPMask surpassed random sampling under both lower and higher masking ratios, indicating that DPPMask makes the reconstruction task more reasonable. We further test our method on the background challenge and multi-class classification tasks, showing that our method is more robust at various tasks.
RepMode: Learning to Re-parameterize Diverse Experts for Subcellular Structure Prediction
Dec 20, 2022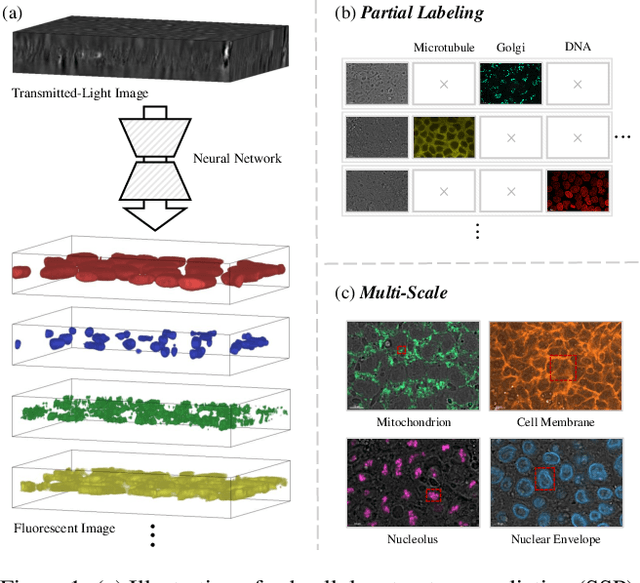
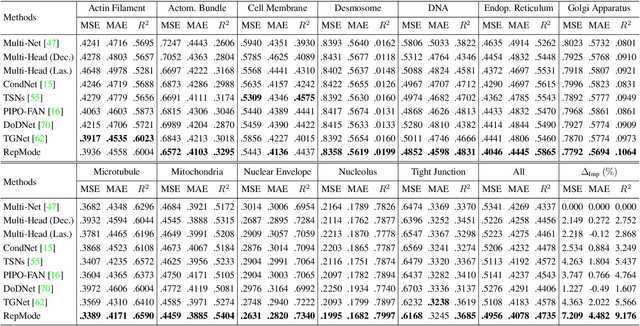
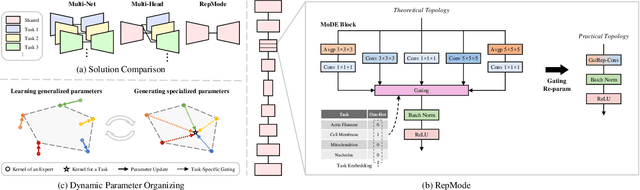
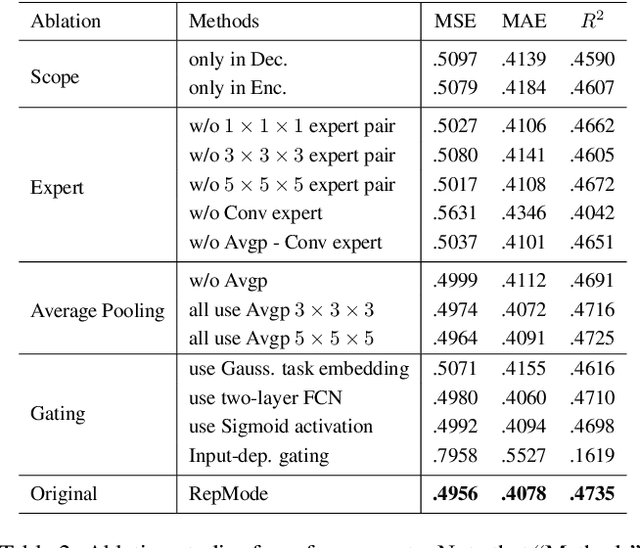
Abstract:In subcellular biological research, fluorescence staining is a key technique to reveal the locations and morphology of subcellular structures. However, fluorescence staining is slow, expensive, and harmful to cells. In this paper, we treat it as a deep learning task termed subcellular structure prediction (SSP), aiming to predict the 3D fluorescent images of multiple subcellular structures from a 3D transmitted-light image. Unfortunately, due to the limitations of current biotechnology, each image is partially labeled in SSP. Besides, naturally, the subcellular structures vary considerably in size, which causes the multi-scale issue in SSP. However, traditional solutions can not address SSP well since they organize network parameters inefficiently and inflexibly. To overcome these challenges, we propose Re-parameterizing Mixture-of-Diverse-Experts (RepMode), a network that dynamically organizes its parameters with task-aware priors to handle specified single-label prediction tasks of SSP. In RepMode, the Mixture-of-Diverse-Experts (MoDE) block is designed to learn the generalized parameters for all tasks, and gating re-parameterization (GatRep) is performed to generate the specialized parameters for each task, by which RepMode can maintain a compact practical topology exactly like a plain network, and meanwhile achieves a powerful theoretical topology. Comprehensive experiments show that RepMode outperforms existing methods on ten of twelve prediction tasks of SSP and achieves state-of-the-art overall performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge