Chunbin Gu
SegRap2025: A Benchmark of Gross Tumor Volume and Lymph Node Clinical Target Volume Segmentation for Radiotherapy Planning of Nasopharyngeal Carcinoma
Jan 28, 2026Abstract:Accurate delineation of Gross Tumor Volume (GTV), Lymph Node Clinical Target Volume (LN CTV), and Organ-at-Risk (OAR) from Computed Tomography (CT) scans is essential for precise radiotherapy planning in Nasopharyngeal Carcinoma (NPC). Building upon SegRap2023, which focused on OAR and GTV segmentation using single-center paired non-contrast CT (ncCT) and contrast-enhanced CT (ceCT) scans, the SegRap2025 challenge aims to enhance the generalizability and robustness of segmentation models across imaging centers and modalities. SegRap2025 comprises two tasks: Task01 addresses GTV segmentation using paired CT from the SegRap2023 dataset, with an additional external testing set to evaluate cross-center generalization, and Task02 focuses on LN CTV segmentation using multi-center training data and an unseen external testing set, where each case contains paired CT scans or a single modality, emphasizing both cross-center and cross-modality robustness. This paper presents the challenge setup and provides a comprehensive analysis of the solutions submitted by ten participating teams. For GTV segmentation task, the top-performing models achieved average Dice Similarity Coefficient (DSC) of 74.61% and 56.79% on the internal and external testing cohorts, respectively. For LN CTV segmentation task, the highest average DSC values reached 60.24%, 60.50%, and 57.23% on paired CT, ceCT-only, and ncCT-only subsets, respectively. SegRap2025 establishes a large-scale multi-center, multi-modality benchmark for evaluating the generalization and robustness in radiotherapy target segmentation, providing valuable insights toward clinically applicable automated radiotherapy planning systems. The benchmark is available at: https://hilab-git.github.io/SegRap2025_Challenge.
Lost in Tokenization: Context as the Key to Unlocking Biomolecular Understanding in Scientific LLMs
Oct 27, 2025Abstract:Scientific Large Language Models (Sci-LLMs) have emerged as a promising frontier for accelerating biological discovery. However, these models face a fundamental challenge when processing raw biomolecular sequences: the tokenization dilemma. Whether treating sequences as a specialized language, risking the loss of functional motif information, or as a separate modality, introducing formidable alignment challenges, current strategies fundamentally limit their reasoning capacity. We challenge this sequence-centric paradigm by positing that a more effective strategy is to provide Sci-LLMs with high-level structured context derived from established bioinformatics tools, thereby bypassing the need to interpret low-level noisy sequence data directly. Through a systematic comparison of leading Sci-LLMs on biological reasoning tasks, we tested three input modes: sequence-only, context-only, and a combination of both. Our findings are striking: the context-only approach consistently and substantially outperforms all other modes. Even more revealing, the inclusion of the raw sequence alongside its high-level context consistently degrades performance, indicating that raw sequences act as informational noise, even for models with specialized tokenization schemes. These results suggest that the primary strength of existing Sci-LLMs lies not in their nascent ability to interpret biomolecular syntax from scratch, but in their profound capacity for reasoning over structured, human-readable knowledge. Therefore, we argue for reframing Sci-LLMs not as sequence decoders, but as powerful reasoning engines over expert knowledge. This work lays the foundation for a new class of hybrid scientific AI agents, repositioning the developmental focus from direct sequence interpretation towards high-level knowledge synthesis. The code is available at github.com/opendatalab-raise-dev/CoKE.
DEL-Ranking: Ranking-Correction Denoising Framework for Elucidating Molecular Affinities in DNA-Encoded Libraries
Oct 19, 2024



Abstract:DNA-encoded library (DEL) screening has revolutionized the detection of protein-ligand interactions through read counts, enabling rapid exploration of vast chemical spaces. However, noise in read counts, stemming from nonspecific interactions, can mislead this exploration process. We present DEL-Ranking, a novel distribution-correction denoising framework that addresses these challenges. Our approach introduces two key innovations: (1) a novel ranking loss that rectifies relative magnitude relationships between read counts, enabling the learning of causal features determining activity levels, and (2) an iterative algorithm employing self-training and consistency loss to establish model coherence between activity label and read count predictions. Furthermore, we contribute three new DEL screening datasets, the first to comprehensively include multi-dimensional molecular representations, protein-ligand enrichment values, and their activity labels. These datasets mitigate data scarcity issues in AI-driven DEL screening research. Rigorous evaluation on diverse DEL datasets demonstrates DEL-Ranking's superior performance across multiple correlation metrics, with significant improvements in binding affinity prediction accuracy. Our model exhibits zero-shot generalization ability across different protein targets and successfully identifies potential motifs determining compound binding affinity. This work advances DEL screening analysis and provides valuable resources for future research in this area.
Unlocking Potential Binders: Multimodal Pretraining DEL-Fusion for Denoising DNA-Encoded Libraries
Sep 07, 2024



Abstract:In the realm of drug discovery, DNA-encoded library (DEL) screening technology has emerged as an efficient method for identifying high-affinity compounds. However, DEL screening faces a significant challenge: noise arising from nonspecific interactions within complex biological systems. Neural networks trained on DEL libraries have been employed to extract compound features, aiming to denoise the data and uncover potential binders to the desired therapeutic target. Nevertheless, the inherent structure of DEL, constrained by the limited diversity of building blocks, impacts the performance of compound encoders. Moreover, existing methods only capture compound features at a single level, further limiting the effectiveness of the denoising strategy. To mitigate these issues, we propose a Multimodal Pretraining DEL-Fusion model (MPDF) that enhances encoder capabilities through pretraining and integrates compound features across various scales. We develop pretraining tasks applying contrastive objectives between different compound representations and their text descriptions, enhancing the compound encoders' ability to acquire generic features. Furthermore, we propose a novel DEL-fusion framework that amalgamates compound information at the atomic, submolecular, and molecular levels, as captured by various compound encoders. The synergy of these innovations equips MPDF with enriched, multi-scale features, enabling comprehensive downstream denoising. Evaluated on three DEL datasets, MPDF demonstrates superior performance in data processing and analysis for validation tasks. Notably, MPDF offers novel insights into identifying high-affinity molecules, paving the way for improved DEL utility in drug discovery.
RepMode: Learning to Re-parameterize Diverse Experts for Subcellular Structure Prediction
Dec 20, 2022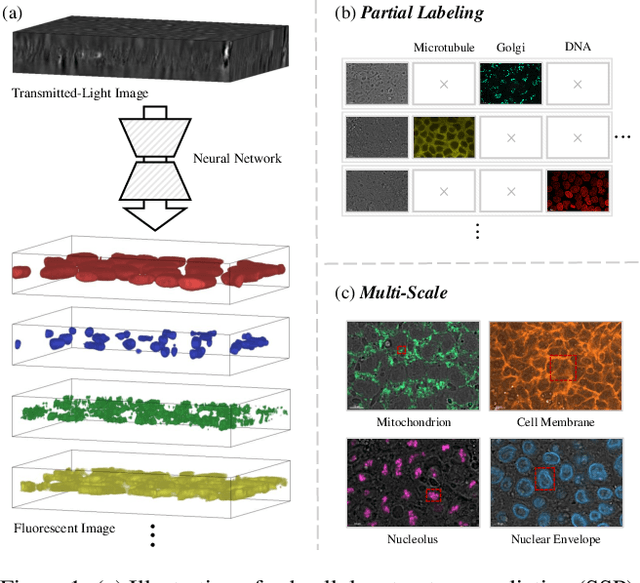
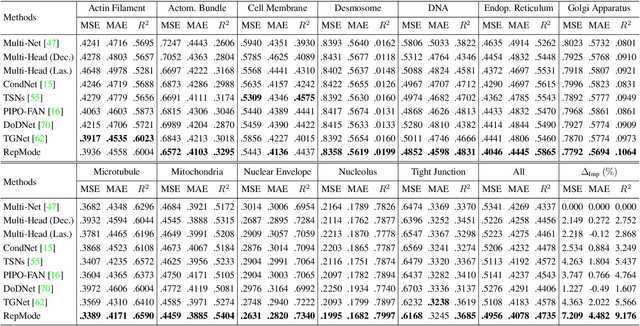
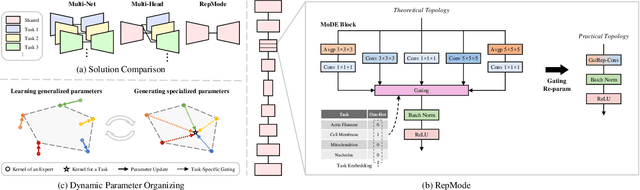
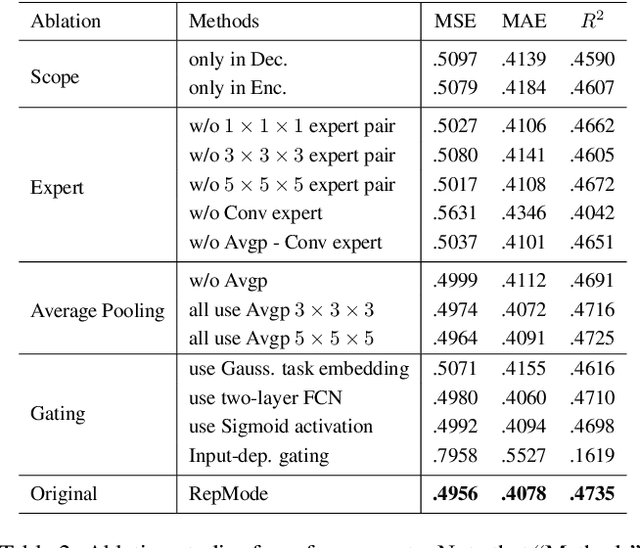
Abstract:In subcellular biological research, fluorescence staining is a key technique to reveal the locations and morphology of subcellular structures. However, fluorescence staining is slow, expensive, and harmful to cells. In this paper, we treat it as a deep learning task termed subcellular structure prediction (SSP), aiming to predict the 3D fluorescent images of multiple subcellular structures from a 3D transmitted-light image. Unfortunately, due to the limitations of current biotechnology, each image is partially labeled in SSP. Besides, naturally, the subcellular structures vary considerably in size, which causes the multi-scale issue in SSP. However, traditional solutions can not address SSP well since they organize network parameters inefficiently and inflexibly. To overcome these challenges, we propose Re-parameterizing Mixture-of-Diverse-Experts (RepMode), a network that dynamically organizes its parameters with task-aware priors to handle specified single-label prediction tasks of SSP. In RepMode, the Mixture-of-Diverse-Experts (MoDE) block is designed to learn the generalized parameters for all tasks, and gating re-parameterization (GatRep) is performed to generate the specialized parameters for each task, by which RepMode can maintain a compact practical topology exactly like a plain network, and meanwhile achieves a powerful theoretical topology. Comprehensive experiments show that RepMode outperforms existing methods on ten of twelve prediction tasks of SSP and achieves state-of-the-art overall performance.
Cross-modal Image Retrieval with Deep Mutual Information Maximization
Mar 10, 2021
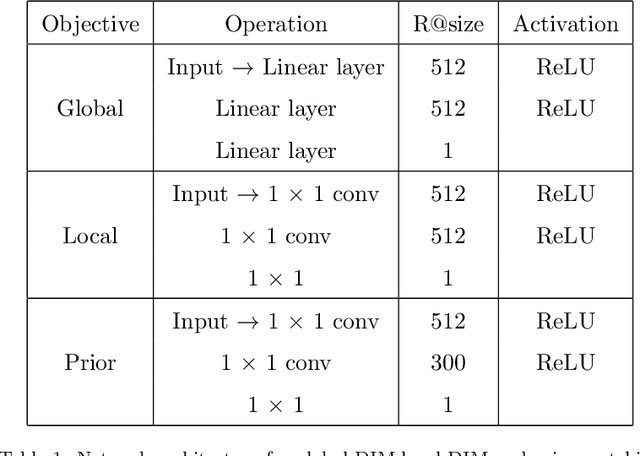
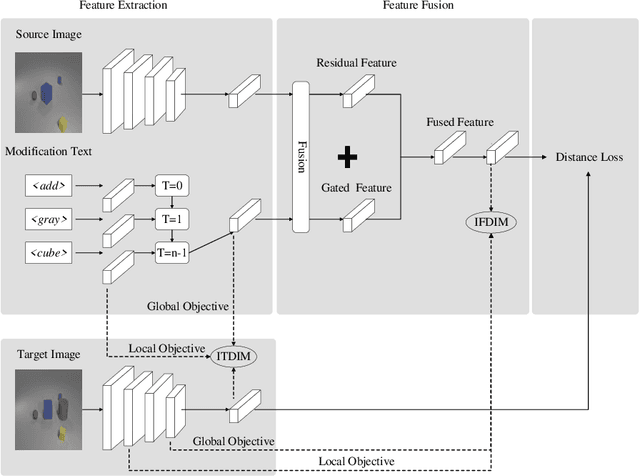
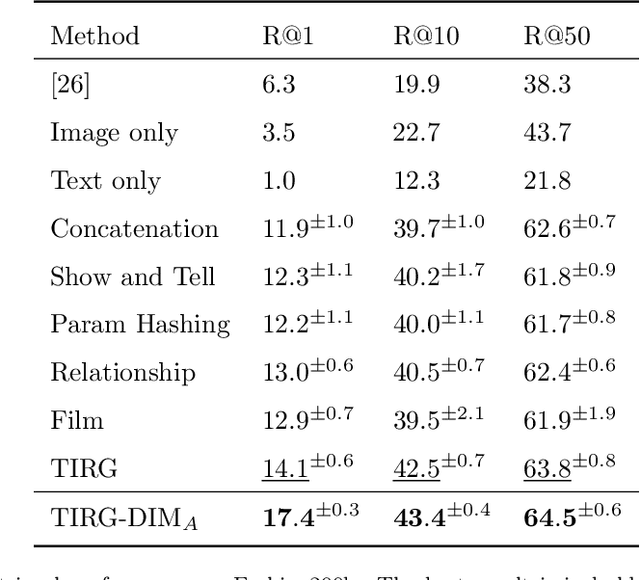
Abstract:In this paper, we study the cross-modal image retrieval, where the inputs contain a source image plus some text that describes certain modifications to this image and the desired image. Prior work usually uses a three-stage strategy to tackle this task: 1) extract the features of the inputs; 2) fuse the feature of the source image and its modified text to obtain fusion feature; 3) learn a similarity metric between the desired image and the source image + modified text by using deep metric learning. Since classical image/text encoders can learn the useful representation and common pair-based loss functions of distance metric learning are enough for cross-modal retrieval, people usually improve retrieval accuracy by designing new fusion networks. However, these methods do not successfully handle the modality gap caused by the inconsistent distribution and representation of the features of different modalities, which greatly influences the feature fusion and similarity learning. To alleviate this problem, we adopt the contrastive self-supervised learning method Deep InforMax (DIM) to our approach to bridge this gap by enhancing the dependence between the text, the image, and their fusion. Specifically, our method narrows the modality gap between the text modality and the image modality by maximizing mutual information between their not exactly semantically identical representation. Moreover, we seek an effective common subspace for the semantically same fusion feature and desired image's feature by utilizing Deep InforMax between the low-level layer of the image encoder and the high-level layer of the fusion network. Extensive experiments on three large-scale benchmark datasets show that we have bridged the modality gap between different modalities and achieve state-of-the-art retrieval performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge