RepMode: Learning to Re-parameterize Diverse Experts for Subcellular Structure Prediction
Paper and Code
Dec 20, 2022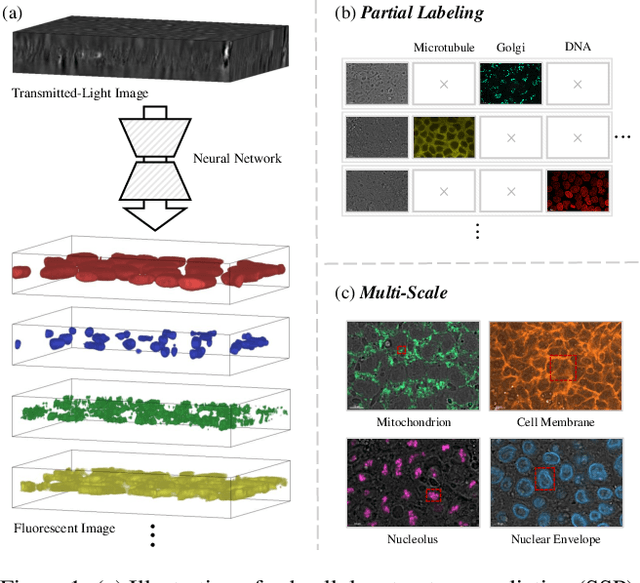
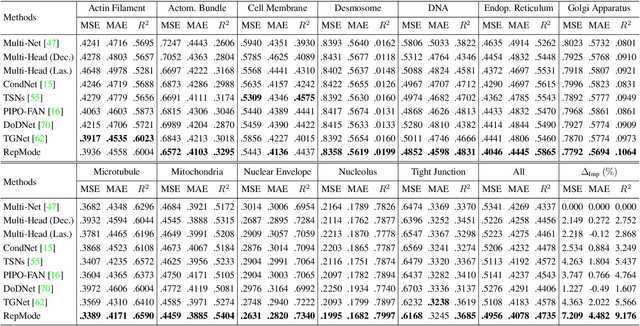
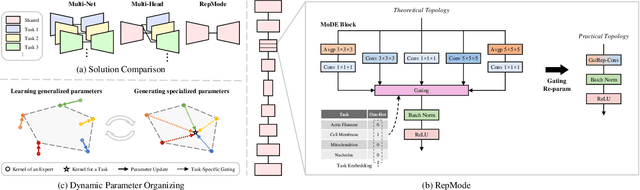
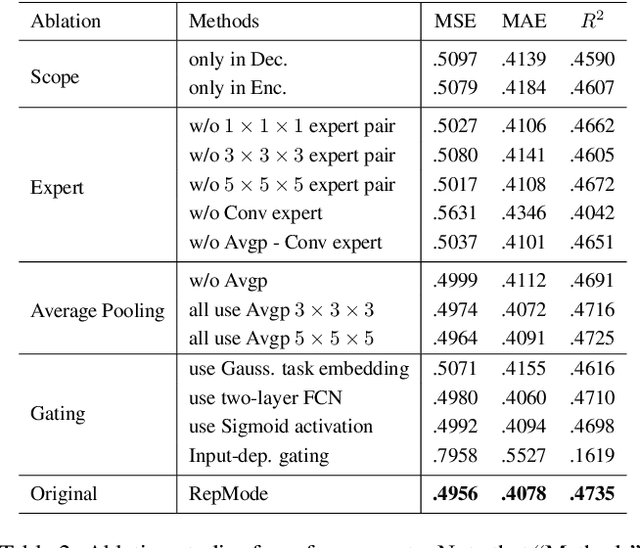
In subcellular biological research, fluorescence staining is a key technique to reveal the locations and morphology of subcellular structures. However, fluorescence staining is slow, expensive, and harmful to cells. In this paper, we treat it as a deep learning task termed subcellular structure prediction (SSP), aiming to predict the 3D fluorescent images of multiple subcellular structures from a 3D transmitted-light image. Unfortunately, due to the limitations of current biotechnology, each image is partially labeled in SSP. Besides, naturally, the subcellular structures vary considerably in size, which causes the multi-scale issue in SSP. However, traditional solutions can not address SSP well since they organize network parameters inefficiently and inflexibly. To overcome these challenges, we propose Re-parameterizing Mixture-of-Diverse-Experts (RepMode), a network that dynamically organizes its parameters with task-aware priors to handle specified single-label prediction tasks of SSP. In RepMode, the Mixture-of-Diverse-Experts (MoDE) block is designed to learn the generalized parameters for all tasks, and gating re-parameterization (GatRep) is performed to generate the specialized parameters for each task, by which RepMode can maintain a compact practical topology exactly like a plain network, and meanwhile achieves a powerful theoretical topology. Comprehensive experiments show that RepMode outperforms existing methods on ten of twelve prediction tasks of SSP and achieves state-of-the-art overall performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge