Xiangyun Liao
SAGS: Self-Adaptive Alias-Free Gaussian Splatting for Dynamic Surgical Endoscopic Reconstruction
Oct 31, 2025Abstract:Surgical reconstruction of dynamic tissues from endoscopic videos is a crucial technology in robot-assisted surgery. The development of Neural Radiance Fields (NeRFs) has greatly advanced deformable tissue reconstruction, achieving high-quality results from video and image sequences. However, reconstructing deformable endoscopic scenes remains challenging due to aliasing and artifacts caused by tissue movement, which can significantly degrade visualization quality. The introduction of 3D Gaussian Splatting (3DGS) has improved reconstruction efficiency by enabling a faster rendering pipeline. Nevertheless, existing 3DGS methods often prioritize rendering speed while neglecting these critical issues. To address these challenges, we propose SAGS, a self-adaptive alias-free Gaussian splatting framework. We introduce an attention-driven, dynamically weighted 4D deformation decoder, leveraging 3D smoothing filters and 2D Mip filters to mitigate artifacts in deformable tissue reconstruction and better capture the fine details of tissue movement. Experimental results on two public benchmarks, EndoNeRF and SCARED, demonstrate that our method achieves superior performance in all metrics of PSNR, SSIM, and LPIPS compared to the state of the art while also delivering better visualization quality.
Versatile and Efficient Medical Image Super-Resolution Via Frequency-Gated Mamba
Oct 31, 2025Abstract:Medical image super-resolution (SR) is essential for enhancing diagnostic accuracy while reducing acquisition cost and scanning time. However, modeling both long-range anatomical structures and fine-grained frequency details with low computational overhead remains challenging. We propose FGMamba, a novel frequency-aware gated state-space model that unifies global dependency modeling and fine-detail enhancement into a lightweight architecture. Our method introduces two key innovations: a Gated Attention-enhanced State-Space Module (GASM) that integrates efficient state-space modeling with dual-branch spatial and channel attention, and a Pyramid Frequency Fusion Module (PFFM) that captures high-frequency details across multiple resolutions via FFT-guided fusion. Extensive evaluations across five medical imaging modalities (Ultrasound, OCT, MRI, CT, and Endoscopic) demonstrate that FGMamba achieves superior PSNR/SSIM while maintaining a compact parameter footprint ($<$0.75M), outperforming CNN-based and Transformer-based SOTAs. Our results validate the effectiveness of frequency-aware state-space modeling for scalable and accurate medical image enhancement.
Progressive Frequency-Aware Network for Laparoscopic Image Desmoking
Dec 19, 2023



Abstract:Laparoscopic surgery offers minimally invasive procedures with better patient outcomes, but smoke presence challenges visibility and safety. Existing learning-based methods demand large datasets and high computational resources. We propose the Progressive Frequency-Aware Network (PFAN), a lightweight GAN framework for laparoscopic image desmoking, combining the strengths of CNN and Transformer for progressive information extraction in the frequency domain. PFAN features CNN-based Multi-scale Bottleneck-Inverting (MBI) Blocks for capturing local high-frequency information and Locally-Enhanced Axial Attention Transformers (LAT) for efficiently handling global low-frequency information. PFAN efficiently desmokes laparoscopic images even with limited training data. Our method outperforms state-of-the-art approaches in PSNR, SSIM, CIEDE2000, and visual quality on the Cholec80 dataset and retains only 629K parameters. Our code and models are made publicly available at: https://github.com/jlzcode/PFAN.
DPPMask: Masked Image Modeling with Determinantal Point Processes
Mar 25, 2023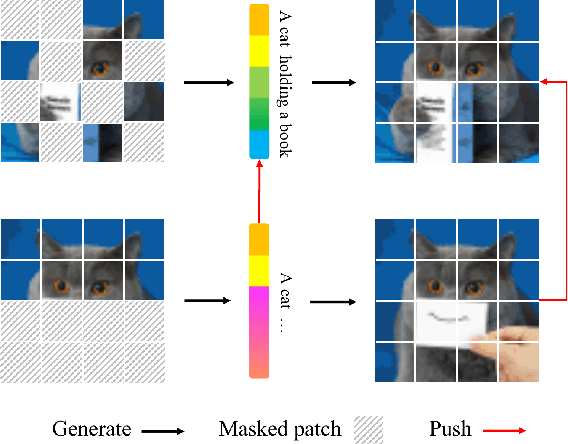
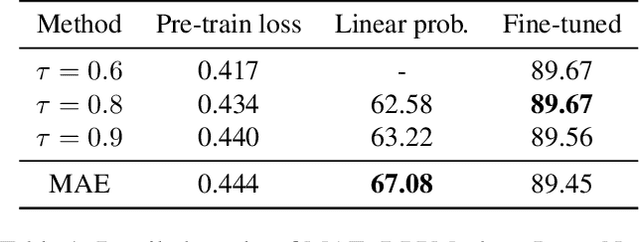
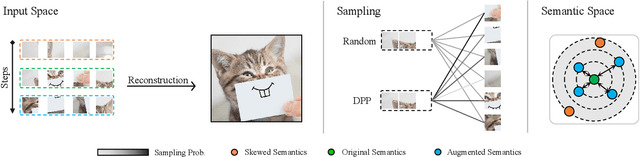
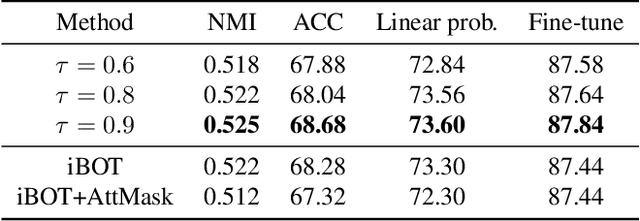
Abstract:Masked Image Modeling (MIM) has achieved impressive representative performance with the aim of reconstructing randomly masked images. Despite the empirical success, most previous works have neglected the important fact that it is unreasonable to force the model to reconstruct something beyond recovery, such as those masked objects. In this work, we show that uniformly random masking widely used in previous works unavoidably loses some key objects and changes original semantic information, resulting in a misalignment problem and hurting the representative learning eventually. To address this issue, we augment MIM with a new masking strategy namely the DPPMask by substituting the random process with Determinantal Point Process (DPPs) to reduce the semantic change of the image after masking. Our method is simple yet effective and requires no extra learnable parameters when implemented within various frameworks. In particular, we evaluate our method on two representative MIM frameworks, MAE and iBOT. We show that DPPMask surpassed random sampling under both lower and higher masking ratios, indicating that DPPMask makes the reconstruction task more reasonable. We further test our method on the background challenge and multi-class classification tasks, showing that our method is more robust at various tasks.
Hepatic vessel segmentation based on 3D swin-transformer with inductive biased multi-head self-attention
Nov 22, 2021



Abstract:Purpose: Segmentation of liver vessels from CT images is indispensable prior to surgical planning and aroused broad range of interests in the medical image analysis community. Due to the complex structure and low contrast background, automatic liver vessel segmentation remains particularly challenging. Most of the related researches adopt FCN, U-net, and V-net variants as a backbone. However, these methods mainly focus on capturing multi-scale local features which may produce misclassified voxels due to the convolutional operator's limited locality reception field. Methods: We propose a robust end-to-end vessel segmentation network called Inductive BIased Multi-Head Attention Vessel Net(IBIMHAV-Net) by expanding swin transformer to 3D and employing an effective combination of convolution and self-attention. In practice, we introduce the voxel-wise embedding rather than patch-wise embedding to locate precise liver vessel voxels, and adopt multi-scale convolutional operators to gain local spatial information. On the other hand, we propose the inductive biased multi-head self-attention which learns inductive biased relative positional embedding from initialized absolute position embedding. Based on this, we can gain a more reliable query and key matrix. To validate the generalization of our model, we test on samples which have different structural complexity. Results: We conducted experiments on the 3DIRCADb datasets. The average dice and sensitivity of the four tested cases were 74.8% and 77.5%, which exceed results of existing deep learning methods and improved graph cuts method. Conclusion: The proposed model IBIMHAV-Net provides an automatic, accurate 3D liver vessel segmentation with an interleaved architecture that better utilizes both global and local spatial features in CT volumes. It can be further extended for other clinical data.
Evaluation of Algorithms for Multi-Modality Whole Heart Segmentation: An Open-Access Grand Challenge
Feb 21, 2019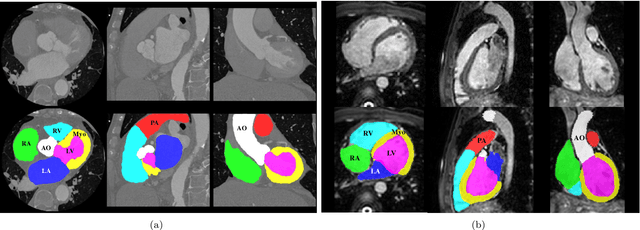
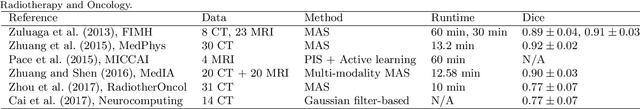
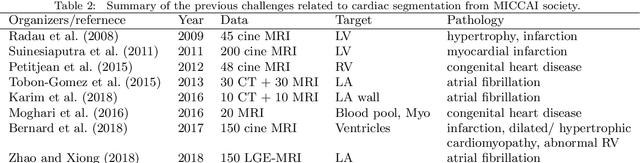
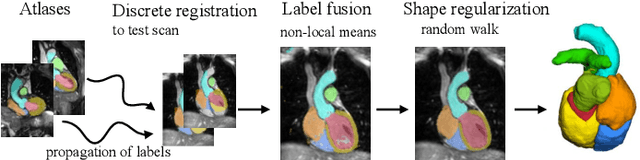
Abstract:Knowledge of whole heart anatomy is a prerequisite for many clinical applications. Whole heart segmentation (WHS), which delineates substructures of the heart, can be very valuable for modeling and analysis of the anatomy and functions of the heart. However, automating this segmentation can be arduous due to the large variation of the heart shape, and different image qualities of the clinical data. To achieve this goal, a set of training data is generally needed for constructing priors or for training. In addition, it is difficult to perform comparisons between different methods, largely due to differences in the datasets and evaluation metrics used. This manuscript presents the methodologies and evaluation results for the WHS algorithms selected from the submissions to the Multi-Modality Whole Heart Segmentation (MM-WHS) challenge, in conjunction with MICCAI 2017. The challenge provides 120 three-dimensional cardiac images covering the whole heart, including 60 CT and 60 MRI volumes, all acquired in clinical environments with manual delineation. Ten algorithms for CT data and eleven algorithms for MRI data, submitted from twelve groups, have been evaluated. The results show that many of the deep learning (DL) based methods achieved high accuracy, even though the number of training datasets was limited. A number of them also reported poor results in the blinded evaluation, probably due to overfitting in their training. The conventional algorithms, mainly based on multi-atlas segmentation, demonstrated robust and stable performance, even though the accuracy is not as good as the best DL method in CT segmentation. The challenge, including the provision of the annotated training data and the blinded evaluation for submitted algorithms on the test data, continues as an ongoing benchmarking resource via its homepage (\url{www.sdspeople.fudan.edu.cn/zhuangxiahai/0/mmwhs/}).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge