Jiahao Huang
Med-CoReasoner: Reducing Language Disparities in Medical Reasoning via Language-Informed Co-Reasoning
Jan 13, 2026Abstract:While reasoning-enhanced large language models perform strongly on English medical tasks, a persistent multilingual gap remains, with substantially weaker reasoning in local languages, limiting equitable global medical deployment. To bridge this gap, we introduce Med-CoReasoner, a language-informed co-reasoning framework that elicits parallel English and local-language reasoning, abstracts them into structured concepts, and integrates local clinical knowledge into an English logical scaffold via concept-level alignment and retrieval. This design combines the structural robustness of English reasoning with the practice-grounded expertise encoded in local languages. To evaluate multilingual medical reasoning beyond multiple-choice settings, we construct MultiMed-X, a benchmark covering seven languages with expert-annotated long-form question answering and natural language inference tasks, comprising 350 instances per language. Experiments across three benchmarks show that Med-CoReasoner improves multilingual reasoning performance by an average of 5%, with particularly substantial gains in low-resource languages. Moreover, model distillation and expert evaluation analysis further confirm that Med-CoReasoner produces clinically sound and culturally grounded reasoning traces.
Convergent and divergent connectivity patterns of the arcuate fasciculus in macaques and humans
Jun 24, 2025Abstract:The organization and connectivity of the arcuate fasciculus (AF) in nonhuman primates remain contentious, especially concerning how its anatomy diverges from that of humans. Here, we combined cross-scale single-neuron tracing - using viral-based genetic labeling and fluorescence micro-optical sectioning tomography in macaques (n = 4; age 3 - 11 years) - with whole-brain tractography from 11.7T diffusion MRI. Complemented by spectral embedding analysis of 7.0T MRI in humans, we performed a comparative connectomic analysis of the AF across species. We demonstrate that the macaque AF originates in the temporal-parietal cortex, traverses the auditory cortex and parietal operculum, and projects into prefrontal regions. In contrast, the human AF exhibits greater expansion into the middle temporal gyrus and stronger prefrontal and parietal operculum connectivity - divergences quantified by Kullback-Leibler analysis that likely underpin the evolutionary specialization of human language networks. These interspecies differences - particularly the human AF's broader temporal integration and strengthened frontoparietal linkages - suggest a connectivity-based substrate for the emergence of advanced language processing unique to humans. Furthermore, our findings offer a neuroanatomical framework for understanding AF-related disorders such as aphasia and dyslexia, where aberrant connectivity disrupts language function.
Topology-Assisted Spatio-Temporal Pattern Disentangling for Scalable MARL in Large-scale Autonomous Traffic Control
Jun 14, 2025Abstract:Intelligent Transportation Systems (ITSs) have emerged as a promising solution towards ameliorating urban traffic congestion, with Traffic Signal Control (TSC) identified as a critical component. Although Multi-Agent Reinforcement Learning (MARL) algorithms have shown potential in optimizing TSC through real-time decision-making, their scalability and effectiveness often suffer from large-scale and complex environments. Typically, these limitations primarily stem from a fundamental mismatch between the exponential growth of the state space driven by the environmental heterogeneities and the limited modeling capacity of current solutions. To address these issues, this paper introduces a novel MARL framework that integrates Dynamic Graph Neural Networks (DGNNs) and Topological Data Analysis (TDA), aiming to enhance the expressiveness of environmental representations and improve agent coordination. Furthermore, inspired by the Mixture of Experts (MoE) architecture in Large Language Models (LLMs), a topology-assisted spatial pattern disentangling (TSD)-enhanced MoE is proposed, which leverages topological signatures to decouple graph features for specialized processing, thus improving the model's ability to characterize dynamic and heterogeneous local observations. The TSD module is also integrated into the policy and value networks of the Multi-agent Proximal Policy Optimization (MAPPO) algorithm, further improving decision-making efficiency and robustness. Extensive experiments conducted on real-world traffic scenarios, together with comprehensive theoretical analysis, validate the superior performance of the proposed framework, highlighting the model's scalability and effectiveness in addressing the complexities of large-scale TSC tasks.
Dynamic Dual Buffer with Divide-and-Conquer Strategy for Online Continual Learning
May 23, 2025Abstract:Online Continual Learning (OCL) presents a complex learning environment in which new data arrives in a batch-to-batch online format, and the risk of catastrophic forgetting can significantly impair model efficacy. In this study, we address OCL by introducing an innovative memory framework that incorporates a short-term memory system to retain dynamic information and a long-term memory system to archive enduring knowledge. Specifically, the long-term memory system comprises a collection of sub-memory buffers, each linked to a cluster prototype and designed to retain data samples from distinct categories. We propose a novel $K$-means-based sample selection method to identify cluster prototypes for each encountered category. To safeguard essential and critical samples, we introduce a novel memory optimisation strategy that selectively retains samples in the appropriate sub-memory buffer by evaluating each cluster prototype against incoming samples through an optimal transportation mechanism. This approach specifically promotes each sub-memory buffer to retain data samples that exhibit significant discrepancies from the corresponding cluster prototype, thereby ensuring the preservation of semantically rich information. In addition, we propose a novel Divide-and-Conquer (DAC) approach that formulates the memory updating as an optimisation problem and divides it into several subproblems. As a result, the proposed DAC approach can solve these subproblems separately and thus can significantly reduce computations of the proposed memory updating process. We conduct a series of experiments across standard and imbalanced learning settings, and the empirical findings indicate that the proposed memory framework achieves state-of-the-art performance in both learning contexts.
Enhancing Diffusion-Weighted Images (DWI) for Diffusion MRI: Is it Enough without Non-Diffusion-Weighted B=0 Reference?
May 19, 2025Abstract:Diffusion MRI (dMRI) is essential for studying brain microstructure, but high-resolution imaging remains challenging due to the inherent trade-offs between acquisition time and signal-to-noise ratio (SNR). Conventional methods often optimize only the diffusion-weighted images (DWIs) without considering their relationship with the non-diffusion-weighted (b=0) reference images. However, calculating diffusion metrics, such as the apparent diffusion coefficient (ADC) and diffusion tensor with its derived metrics like fractional anisotropy (FA) and mean diffusivity (MD), relies on the ratio between each DWI and the b=0 image, which is crucial for clinical observation and diagnostics. In this study, we demonstrate that solely enhancing DWIs using a conventional pixel-wise mean squared error (MSE) loss is insufficient, as the error in ratio between generated DWIs and b=0 diverges. We propose a novel ratio loss, defined as the MSE loss between the predicted and ground-truth log of DWI/b=0 ratios. Our results show that incorporating the ratio loss significantly improves the convergence of this ratio error, achieving lower ratio MSE and slightly enhancing the peak signal-to-noise ratio (PSNR) of generated DWIs. This leads to improved dMRI super-resolution and better preservation of b=0 ratio-based features for the derivation of diffusion metrics.
Decoupling Multi-Contrast Super-Resolution: Pairing Unpaired Synthesis with Implicit Representations
May 09, 2025Abstract:Magnetic Resonance Imaging (MRI) is critical for clinical diagnostics but is often limited by long acquisition times and low signal-to-noise ratios, especially in modalities like diffusion and functional MRI. The multi-contrast nature of MRI presents a valuable opportunity for cross-modal enhancement, where high-resolution (HR) modalities can serve as references to boost the quality of their low-resolution (LR) counterparts-motivating the development of Multi-Contrast Super-Resolution (MCSR) techniques. Prior work has shown that leveraging complementary contrasts can improve SR performance; however, effective feature extraction and fusion across modalities with varying resolutions remains a major challenge. Moreover, existing MCSR methods often assume fixed resolution settings and all require large, perfectly paired training datasets-conditions rarely met in real-world clinical environments. To address these challenges, we propose a novel Modular Multi-Contrast Super-Resolution (MCSR) framework that eliminates the need for paired training data and supports arbitrary upscaling. Our method decouples the MCSR task into two stages: (1) Unpaired Cross-Modal Synthesis (U-CMS), which translates a high-resolution reference modality into a synthesized version of the target contrast, and (2) Unsupervised Super-Resolution (U-SR), which reconstructs the final output using implicit neural representations (INRs) conditioned on spatial coordinates. This design enables scale-agnostic and anatomically faithful reconstruction by bridging un-paired cross-modal synthesis with unsupervised resolution enhancement. Experiments show that our method achieves superior performance at 4x and 8x upscaling, with improved fidelity and anatomical consistency over existing baselines. Our framework demonstrates strong potential for scalable, subject-specific, and data-efficient MCSR in real-world clinical settings.
An Arbitrary-Modal Fusion Network for Volumetric Cranial Nerves Tract Segmentation
May 05, 2025Abstract:The segmentation of cranial nerves (CNs) tract provides a valuable quantitative tool for the analysis of the morphology and trajectory of individual CNs. Multimodal CNs tract segmentation networks, e.g., CNTSeg, which combine structural Magnetic Resonance Imaging (MRI) and diffusion MRI, have achieved promising segmentation performance. However, it is laborious or even infeasible to collect complete multimodal data in clinical practice due to limitations in equipment, user privacy, and working conditions. In this work, we propose a novel arbitrary-modal fusion network for volumetric CNs tract segmentation, called CNTSeg-v2, which trains one model to handle different combinations of available modalities. Instead of directly combining all the modalities, we select T1-weighted (T1w) images as the primary modality due to its simplicity in data acquisition and contribution most to the results, which supervises the information selection of other auxiliary modalities. Our model encompasses an Arbitrary-Modal Collaboration Module (ACM) designed to effectively extract informative features from other auxiliary modalities, guided by the supervision of T1w images. Meanwhile, we construct a Deep Distance-guided Multi-stage (DDM) decoder to correct small errors and discontinuities through signed distance maps to improve segmentation accuracy. We evaluate our CNTSeg-v2 on the Human Connectome Project (HCP) dataset and the clinical Multi-shell Diffusion MRI (MDM) dataset. Extensive experimental results show that our CNTSeg-v2 achieves state-of-the-art segmentation performance, outperforming all competing methods.
RSFR: A Coarse-to-Fine Reconstruction Framework for Diffusion Tensor Cardiac MRI with Semantic-Aware Refinement
Apr 25, 2025Abstract:Cardiac diffusion tensor imaging (DTI) offers unique insights into cardiomyocyte arrangements, bridging the gap between microscopic and macroscopic cardiac function. However, its clinical utility is limited by technical challenges, including a low signal-to-noise ratio, aliasing artefacts, and the need for accurate quantitative fidelity. To address these limitations, we introduce RSFR (Reconstruction, Segmentation, Fusion & Refinement), a novel framework for cardiac diffusion-weighted image reconstruction. RSFR employs a coarse-to-fine strategy, leveraging zero-shot semantic priors via the Segment Anything Model and a robust Vision Mamba-based reconstruction backbone. Our framework integrates semantic features effectively to mitigate artefacts and enhance fidelity, achieving state-of-the-art reconstruction quality and accurate DT parameter estimation under high undersampling rates. Extensive experiments and ablation studies demonstrate the superior performance of RSFR compared to existing methods, highlighting its robustness, scalability, and potential for clinical translation in quantitative cardiac DTI.
LLM-as-a-Judge: Reassessing the Performance of LLMs in Extractive QA
Apr 16, 2025
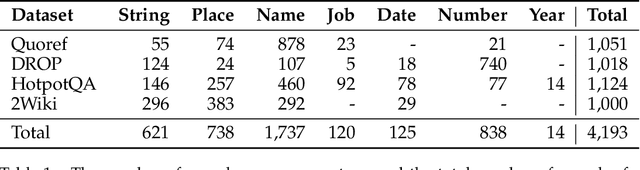
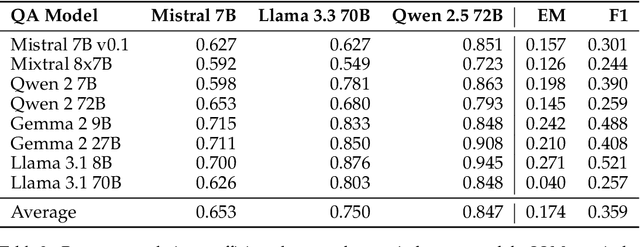
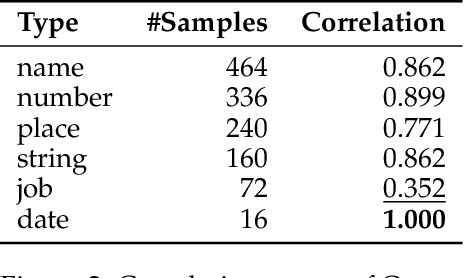
Abstract:Extractive reading comprehension question answering (QA) datasets are typically evaluated using Exact Match (EM) and F1-score, but these metrics often fail to fully capture model performance. With the success of large language models (LLMs), they have been employed in various tasks, including serving as judges (LLM-as-a-judge). In this paper, we reassess the performance of QA models using LLM-as-a-judge across four reading comprehension QA datasets. We examine different families of LLMs and various answer types to evaluate the effectiveness of LLM-as-a-judge in these tasks. Our results show that LLM-as-a-judge is highly correlated with human judgments and can replace traditional EM/F1 metrics. By using LLM-as-a-judge, the correlation with human judgments improves significantly, from 0.17 (EM) and 0.36 (F1-score) to 0.85. These findings confirm that EM and F1 metrics underestimate the true performance of the QA models. While LLM-as-a-judge is not perfect for more difficult answer types (e.g., job), it still outperforms EM/F1, and we observe no bias issues, such as self-preference, when the same model is used for both the QA and judgment tasks.
Is fitting error a reliable metric for assessing deformable motion correction in quantitative MRI?
Mar 10, 2025Abstract:Quantitative MR (qMR) can provide numerical values representing the physical and chemical properties of the tissues. To collect a series of frames under varying settings, retrospective motion correction is essential to align the corresponding anatomical points or features. Under the assumption that the misalignment makes the discrepancy between the corresponding features larger, fitting error is a commonly used evaluation metric for motion correction in qMR. This study evaluates the reliability of the fitting error metric in cardiac diffusion tensor imaging (cDTI) after deformable registration. We found that while fitting error correlates with the negative eigenvalues, the negative Jacobian Determinant increases with broken cardiomyocytes, indicated by helix angle gradient line profiles. Since fitting error measures the distance between moved points and their re-rendered counterparts, the fitting parameter itself may be adjusted due to poor registration. Therefore, fitting error in deformable registration itself is a necessary but not sufficient metric and should be combined with other metrics.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge