Fahim Ahmed Zaman
Denoising Diffusions in Latent Space for Medical Image Segmentation
Jul 17, 2024Abstract:Diffusion models (DPMs) have demonstrated remarkable performance in image generation, often times outperforming other generative models. Since their introduction, the powerful noise-to-image denoising pipeline has been extended to various discriminative tasks, including image segmentation. In case of medical imaging, often times the images are large 3D scans, where segmenting one image using DPMs become extremely inefficient due to large memory consumption and time consuming iterative sampling process. In this work, we propose a novel conditional generative modeling framework (LDSeg) that performs diffusion in latent space for medical image segmentation. Our proposed framework leverages the learned inherent low-dimensional latent distribution of the target object shapes and source image embeddings. The conditional diffusion in latent space not only ensures accurate n-D image segmentation for multi-label objects, but also mitigates the major underlying problems of the traditional DPM based segmentation: (1) large memory consumption, (2) time consuming sampling process and (3) unnatural noise injection in forward/reverse process. LDSeg achieved state-of-the-art segmentation accuracy on three medical image datasets with different imaging modalities. Furthermore, we show that our proposed model is significantly more robust to noises, compared to the traditional deterministic segmentation models, which can be potential in solving the domain shift problems in the medical imaging domain. Codes are available at: https://github.com/LDSeg/LDSeg.
Diagnosis Of Takotsubo Syndrome By Robust Feature Selection From The Complex Latent Space Of DL-based Segmentation Network
Dec 19, 2023Abstract:Researchers have shown significant correlations among segmented objects in various medical imaging modalities and disease related pathologies. Several studies showed that using hand crafted features for disease prediction neglects the immense possibility to use latent features from deep learning (DL) models which may reduce the overall accuracy of differential diagnosis. However, directly using classification or segmentation models on medical to learn latent features opt out robust feature selection and may lead to overfitting. To fill this gap, we propose a novel feature selection technique using the latent space of a segmentation model that can aid diagnosis. We evaluated our method in differentiating a rare cardiac disease: Takotsubo Syndrome (TTS) from the ST elevation myocardial infarction (STEMI) using echocardiogram videos (echo). TTS can mimic clinical features of STEMI in echo and extremely hard to distinguish. Our approach shows promising results in differential diagnosis of TTS with 82% diagnosis accuracy beating the previous state-of-the-art (SOTA) approach. Moreover, the robust feature selection technique using LASSO algorithm shows great potential in reducing the redundant features and creates a robust pipeline for short- and long-term disease prognoses in the downstream analysis.
Surf-CDM: Score-Based Surface Cold-Diffusion Model For Medical Image Segmentation
Dec 19, 2023Abstract:Diffusion models have shown impressive performance for image generation, often times outperforming other generative models. Since their introduction, researchers have extended the powerful noise-to-image denoising pipeline to discriminative tasks, including image segmentation. In this work we propose a conditional score-based generative modeling framework for medical image segmentation which relies on a parametric surface representation for the segmentation masks. The surface re-parameterization allows the direct application of standard diffusion theory, as opposed to when the mask is represented as a binary mask. Moreover, we adapted an extended variant of the diffusion technique known as the "cold-diffusion" where the diffusion model can be constructed with deterministic perturbations instead of Gaussian noise, which facilitates significantly faster convergence in the reverse diffusion. We evaluated our method on the segmentation of the left ventricle from 65 transthoracic echocardiogram videos (2230 echo image frames) and compared its performance to the most popular and widely used image segmentation models. Our proposed model not only outperformed the compared methods in terms of segmentation accuracy, but also showed potential in estimating segmentation uncertainties for further downstream analyses due to its inherent generative nature.
Trust, but Verify: Robust Image Segmentation using Deep Learning
Oct 29, 2023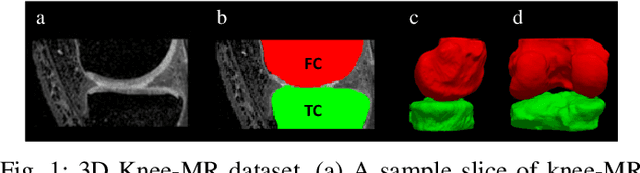
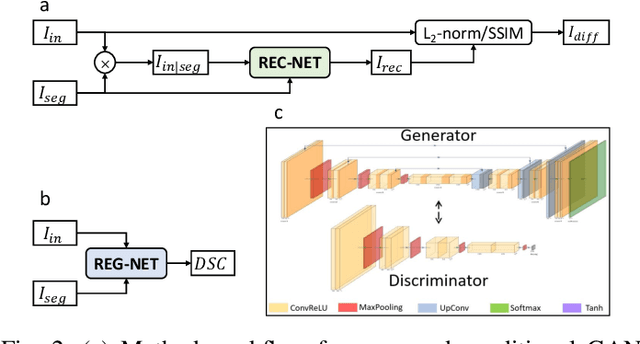
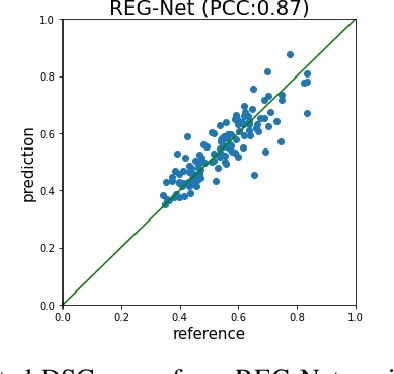
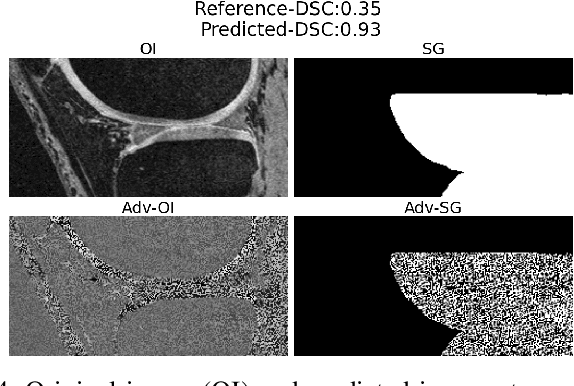
Abstract:We describe a method for verifying the output of a deep neural network for medical image segmentation that is robust to several classes of random as well as worst-case perturbations i.e. adversarial attacks. This method is based on a general approach recently developed by the authors called "Trust, but Verify" wherein an auxiliary verification network produces predictions about certain masked features in the input image using the segmentation as an input. A well-designed auxiliary network will produce high-quality predictions when the input segmentations are accurate, but will produce low-quality predictions when the segmentations are incorrect. Checking the predictions of such a network with the original image allows us to detect bad segmentations. However, to ensure the verification method is truly robust, we need a method for checking the quality of the predictions that does not itself rely on a black-box neural network. Indeed, we show that previous methods for segmentation evaluation that do use deep neural regression networks are vulnerable to false negatives i.e. can inaccurately label bad segmentations as good. We describe the design of a verification network that avoids such vulnerability and present results to demonstrate its robustness compared to previous methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge