Nishchal Sapkota
H-CNN-ViT: A Hierarchical Gated Attention Multi-Branch Model for Bladder Cancer Recurrence Prediction
Nov 19, 2025Abstract:Bladder cancer is one of the most prevalent malignancies worldwide, with a recurrence rate of up to 78%, necessitating accurate post-operative monitoring for effective patient management. Multi-sequence contrast-enhanced MRI is commonly used for recurrence detection; however, interpreting these scans remains challenging, even for experienced radiologists, due to post-surgical alterations such as scarring, swelling, and tissue remodeling. AI-assisted diagnostic tools have shown promise in improving bladder cancer recurrence prediction, yet progress in this field is hindered by the lack of dedicated multi-sequence MRI datasets for recurrence assessment study. In this work, we first introduce a curated multi-sequence, multi-modal MRI dataset specifically designed for bladder cancer recurrence prediction, establishing a valuable benchmark for future research. We then propose H-CNN-ViT, a new Hierarchical Gated Attention Multi-Branch model that enables selective weighting of features from the global (ViT) and local (CNN) paths based on contextual demands, achieving a balanced and targeted feature fusion. Our multi-branch architecture processes each modality independently, ensuring that the unique properties of each imaging channel are optimally captured and integrated. Evaluated on our dataset, H-CNN-ViT achieves an AUC of 78.6%, surpassing state-of-the-art models. Our model is publicly available at https://github.com/XLIAaron/H-CNN-ViT.
When Swin Transformer Meets KANs: An Improved Transformer Architecture for Medical Image Segmentation
Nov 06, 2025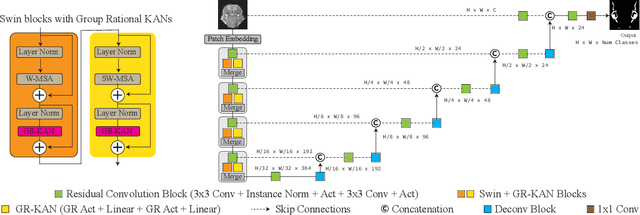
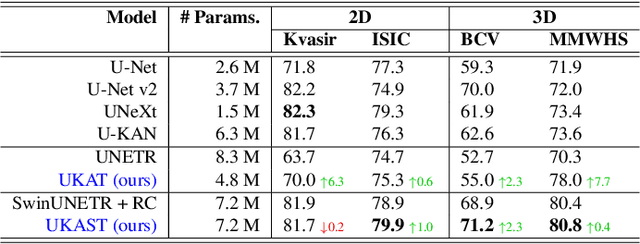
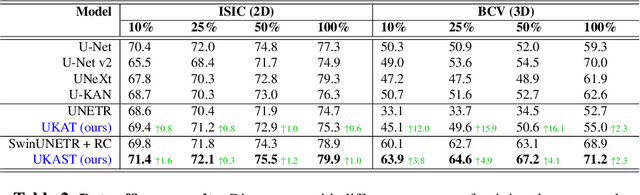
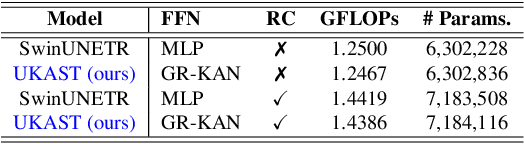
Abstract:Medical image segmentation is critical for accurate diagnostics and treatment planning, but remains challenging due to complex anatomical structures and limited annotated training data. CNN-based segmentation methods excel at local feature extraction, but struggle with modeling long-range dependencies. Transformers, on the other hand, capture global context more effectively, but are inherently data-hungry and computationally expensive. In this work, we introduce UKAST, a U-Net like architecture that integrates rational-function based Kolmogorov-Arnold Networks (KANs) into Swin Transformer encoders. By leveraging rational base functions and Group Rational KANs (GR-KANs) from the Kolmogorov-Arnold Transformer (KAT), our architecture addresses the inefficiencies of vanilla spline-based KANs, yielding a more expressive and data-efficient framework with reduced FLOPs and only a very small increase in parameter count compared to SwinUNETR. UKAST achieves state-of-the-art performance on four diverse 2D and 3D medical image segmentation benchmarks, consistently surpassing both CNN- and Transformer-based baselines. Notably, it attains superior accuracy in data-scarce settings, alleviating the data-hungry limitations of standard Vision Transformers. These results show the potential of KAN-enhanced Transformers to advance data-efficient medical image segmentation. Code is available at: https://github.com/nsapkota417/UKAST
UniCoN: Universal Conditional Networks for Multi-Age Embryonic Cartilage Segmentation with Sparsely Annotated Data
Oct 16, 2024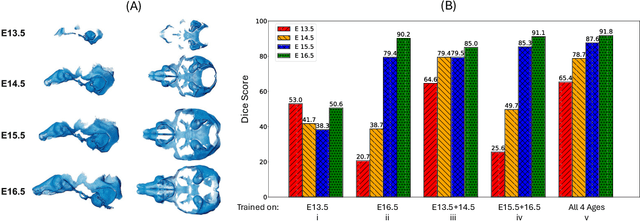
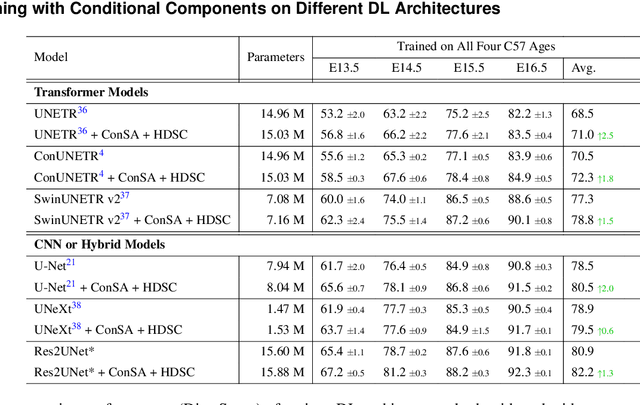
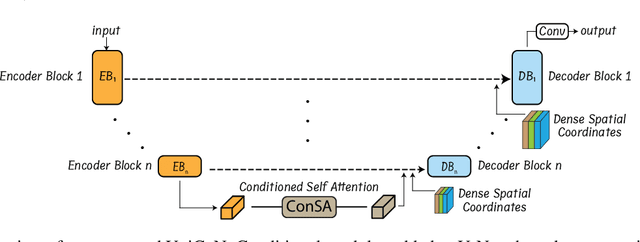
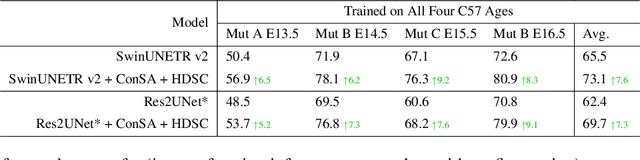
Abstract:Osteochondrodysplasia, affecting 2-3% of newborns globally, is a group of bone and cartilage disorders that often result in head malformations, contributing to childhood morbidity and reduced quality of life. Current research on this disease using mouse models faces challenges since it involves accurately segmenting the developing cartilage in 3D micro-CT images of embryonic mice. Tackling this segmentation task with deep learning (DL) methods is laborious due to the big burden of manual image annotation, expensive due to the high acquisition costs of 3D micro-CT images, and difficult due to embryonic cartilage's complex and rapidly changing shapes. While DL approaches have been proposed to automate cartilage segmentation, most such models have limited accuracy and generalizability, especially across data from different embryonic age groups. To address these limitations, we propose novel DL methods that can be adopted by any DL architectures -- including CNNs, Transformers, or hybrid models -- which effectively leverage age and spatial information to enhance model performance. Specifically, we propose two new mechanisms, one conditioned on discrete age categories and the other on continuous image crop locations, to enable an accurate representation of cartilage shape changes across ages and local shape details throughout the cranial region. Extensive experiments on multi-age cartilage segmentation datasets show significant and consistent performance improvements when integrating our conditional modules into popular DL segmentation architectures. On average, we achieve a 1.7% Dice score increase with minimal computational overhead and a 7.5% improvement on unseen data. These results highlight the potential of our approach for developing robust, universal models capable of handling diverse datasets with limited annotated data, a key challenge in DL-based medical image analysis.
Boosting Medical Image Classification with Segmentation Foundation Model
Jun 16, 2024
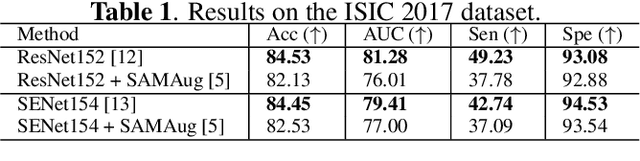
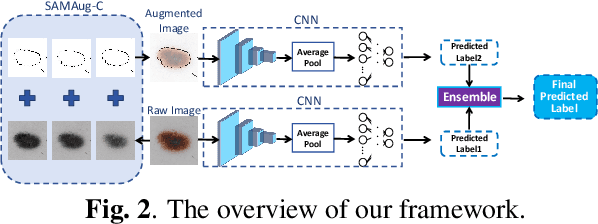
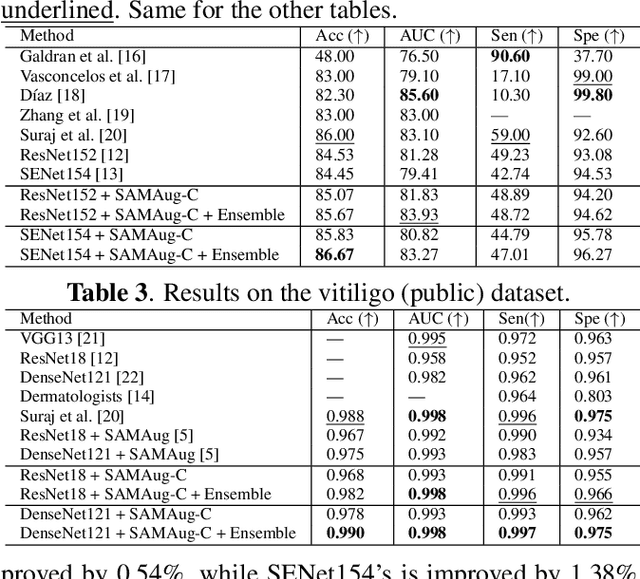
Abstract:The Segment Anything Model (SAM) exhibits impressive capabilities in zero-shot segmentation for natural images. Recently, SAM has gained a great deal of attention for its applications in medical image segmentation. However, to our best knowledge, no studies have shown how to harness the power of SAM for medical image classification. To fill this gap and make SAM a true ``foundation model'' for medical image analysis, it is highly desirable to customize SAM specifically for medical image classification. In this paper, we introduce SAMAug-C, an innovative augmentation method based on SAM for augmenting classification datasets by generating variants of the original images. The augmented datasets can be used to train a deep learning classification model, thereby boosting the classification performance. Furthermore, we propose a novel framework that simultaneously processes raw and SAMAug-C augmented image input, capitalizing on the complementary information that is offered by both. Experiments on three public datasets validate the effectiveness of our new approach.
Path-GPTOmic: A Balanced Multi-modal Learning Framework for Survival Outcome Prediction
Mar 18, 2024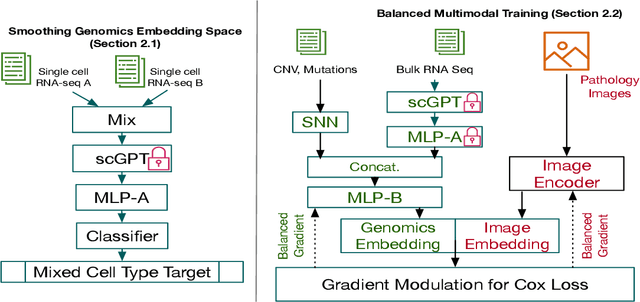
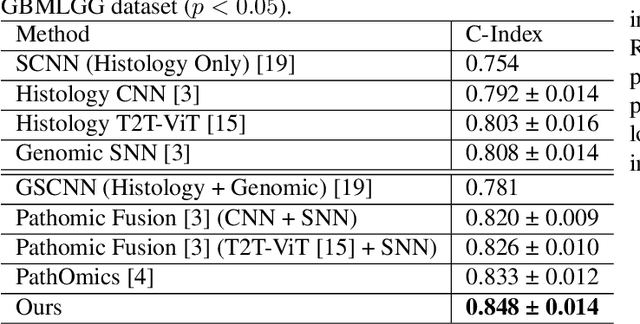

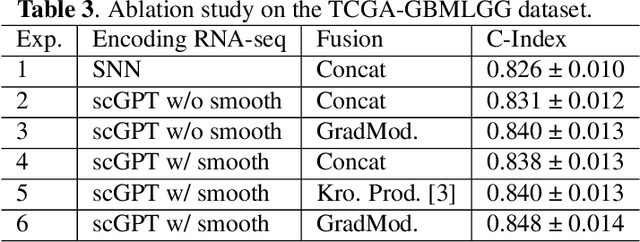
Abstract:For predicting cancer survival outcomes, standard approaches in clinical research are often based on two main modalities: pathology images for observing cell morphology features, and genomic (e.g., bulk RNA-seq) for quantifying gene expressions. However, existing pathology-genomic multi-modal algorithms face significant challenges: (1) Valuable biological insights regarding genes and gene-gene interactions are frequently overlooked; (2) one modality often dominates the optimization process, causing inadequate training for the other modality. In this paper, we introduce a new multi-modal ``Path-GPTOmic" framework for cancer survival outcome prediction. First, to extract valuable biological insights, we regulate the embedding space of a foundation model, scGPT, initially trained on single-cell RNA-seq data, making it adaptable for bulk RNA-seq data. Second, to address the imbalance-between-modalities problem, we propose a gradient modulation mechanism tailored to the Cox partial likelihood loss for survival prediction. The contributions of the modalities are dynamically monitored and adjusted during the training process, encouraging that both modalities are sufficiently trained. Evaluated on two TCGA(The Cancer Genome Atlas) datasets, our model achieves substantially improved survival prediction accuracy.
SHMC-Net: A Mask-guided Feature Fusion Network for Sperm Head Morphology Classification
Feb 07, 2024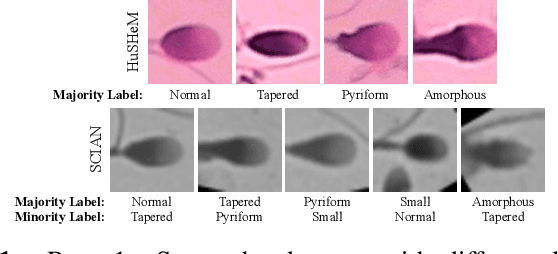
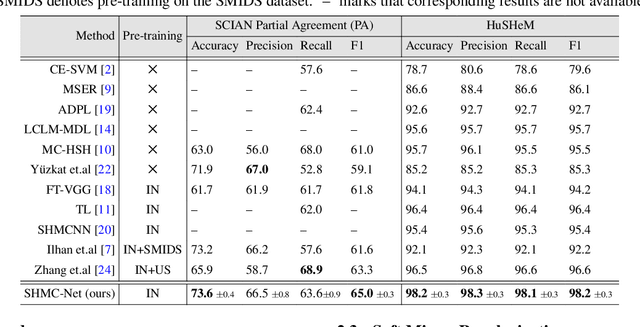
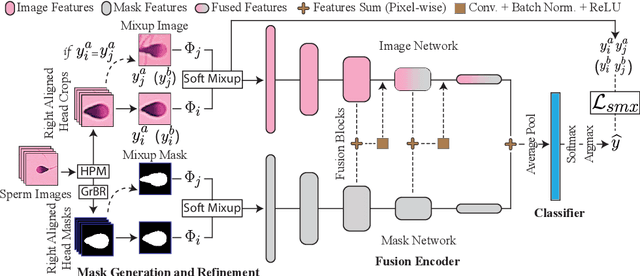
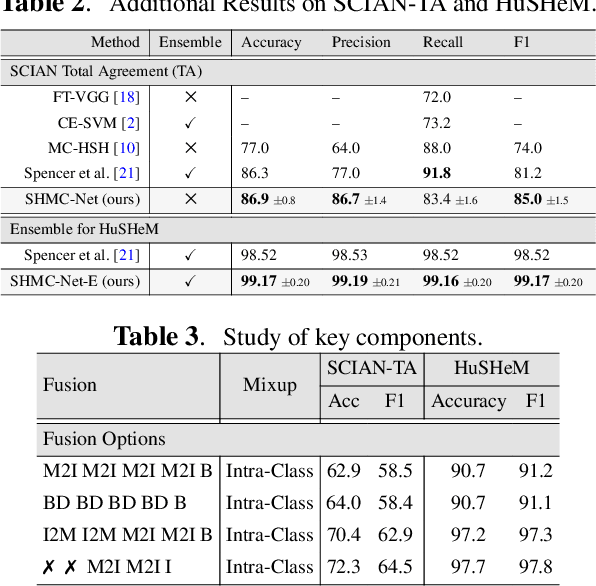
Abstract:Male infertility accounts for about one-third of global infertility cases. Manual assessment of sperm abnormalities through head morphology analysis encounters issues of observer variability and diagnostic discrepancies among experts. Its alternative, Computer-Assisted Semen Analysis (CASA), suffers from low-quality sperm images, small datasets, and noisy class labels. We propose a new approach for sperm head morphology classification, called SHMC-Net, which uses segmentation masks of sperm heads to guide the morphology classification of sperm images. SHMC-Net generates reliable segmentation masks using image priors, refines object boundaries with an efficient graph-based method, and trains an image network with sperm head crops and a mask network with the corresponding masks. In the intermediate stages of the networks, image and mask features are fused with a fusion scheme to better learn morphological features. To handle noisy class labels and regularize training on small datasets, SHMC-Net applies Soft Mixup to combine mixup augmentation and a loss function. We achieve state-of-the-art results on SCIAN and HuSHeM datasets, outperforming methods that use additional pre-training or costly ensembling techniques.
ConUNETR: A Conditional Transformer Network for 3D Micro-CT Embryonic Cartilage Segmentation
Feb 06, 2024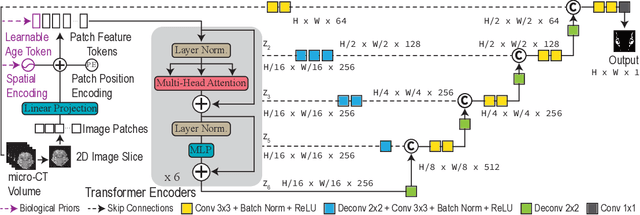
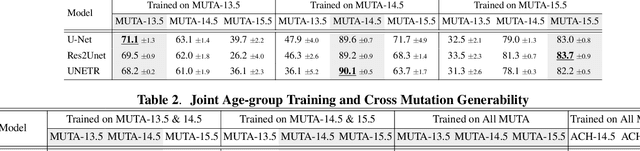
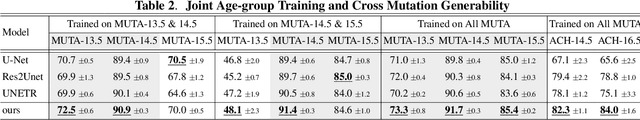
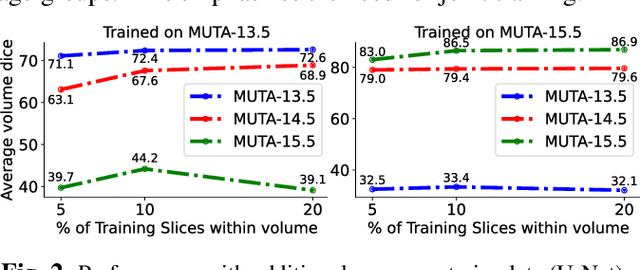
Abstract:Studying the morphological development of cartilaginous and osseous structures is critical to the early detection of life-threatening skeletal dysmorphology. Embryonic cartilage undergoes rapid structural changes within hours, introducing biological variations and morphological shifts that limit the generalization of deep learning-based segmentation models that infer across multiple embryonic age groups. Obtaining individual models for each age group is expensive and less effective, while direct transfer (predicting an age unseen during training) suffers a potential performance drop due to morphological shifts. We propose a novel Transformer-based segmentation model with improved biological priors that better distills morphologically diverse information through conditional mechanisms. This enables a single model to accurately predict cartilage across multiple age groups. Experiments on the mice cartilage dataset show the superiority of our new model compared to other competitive segmentation models. Additional studies on a separate mice cartilage dataset with a distinct mutation show that our model generalizes well and effectively captures age-based cartilage morphology patterns.
SwIPE: Efficient and Robust Medical Image Segmentation with Implicit Patch Embeddings
Jul 23, 2023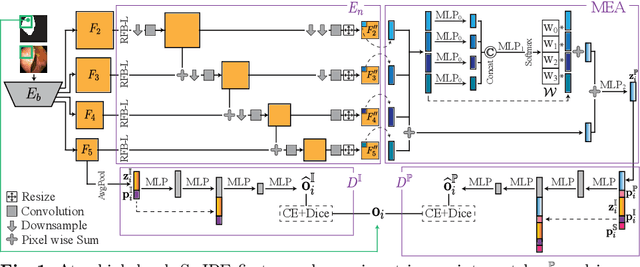
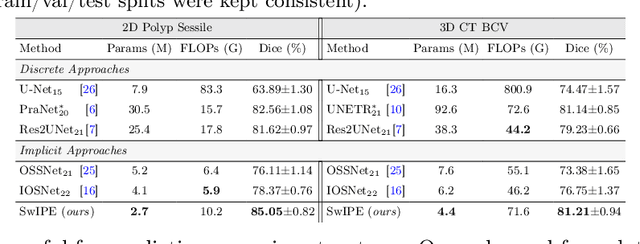

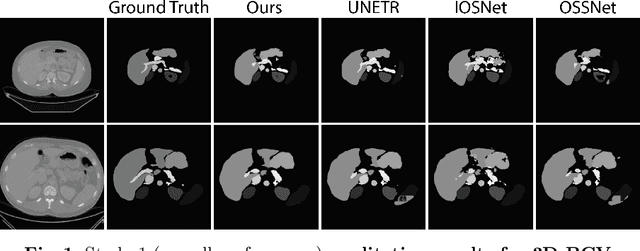
Abstract:Modern medical image segmentation methods primarily use discrete representations in the form of rasterized masks to learn features and generate predictions. Although effective, this paradigm is spatially inflexible, scales poorly to higher-resolution images, and lacks direct understanding of object shapes. To address these limitations, some recent works utilized implicit neural representations (INRs) to learn continuous representations for segmentation. However, these methods often directly adopted components designed for 3D shape reconstruction. More importantly, these formulations were also constrained to either point-based or global contexts, lacking contextual understanding or local fine-grained details, respectively--both critical for accurate segmentation. To remedy this, we propose a novel approach, SwIPE (Segmentation with Implicit Patch Embeddings), that leverages the advantages of INRs and predicts shapes at the patch level--rather than at the point level or image level--to enable both accurate local boundary delineation and global shape coherence. Extensive evaluations on two tasks (2D polyp segmentation and 3D abdominal organ segmentation) show that SwIPE significantly improves over recent implicit approaches and outperforms state-of-the-art discrete methods with over 10x fewer parameters. Our method also demonstrates superior data efficiency and improved robustness to data shifts across image resolutions and datasets. Code is available on Github.
Keep Your Friends Close & Enemies Farther: Debiasing Contrastive Learning with Spatial Priors in 3D Radiology Images
Nov 16, 2022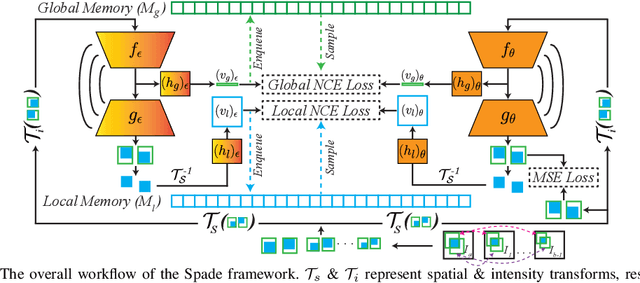
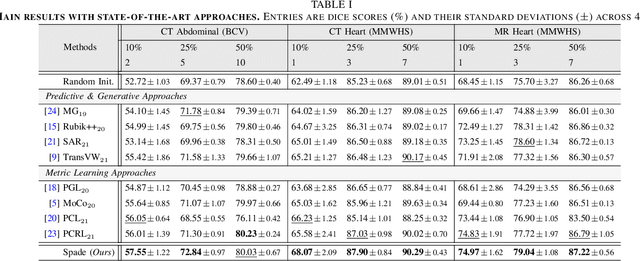
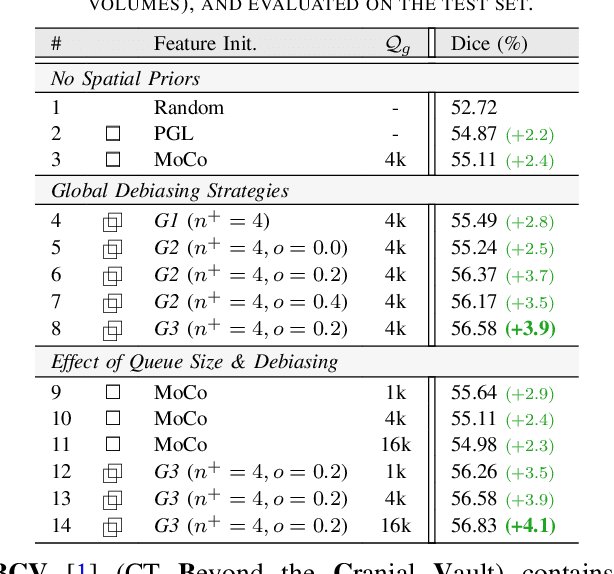
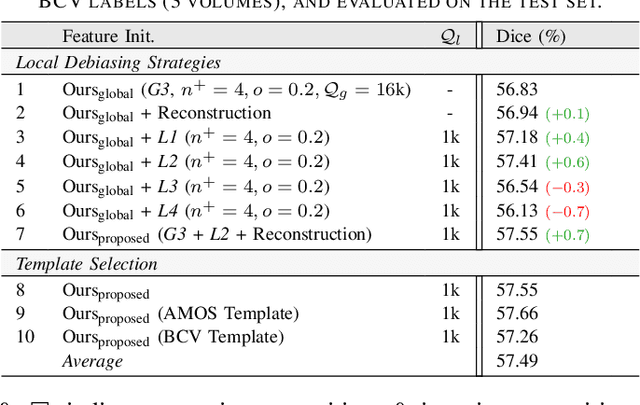
Abstract:Understanding of spatial attributes is central to effective 3D radiology image analysis where crop-based learning is the de facto standard. Given an image patch, its core spatial properties (e.g., position & orientation) provide helpful priors on expected object sizes, appearances, and structures through inherent anatomical consistencies. Spatial correspondences, in particular, can effectively gauge semantic similarities between inter-image regions, while their approximate extraction requires no annotations or overbearing computational costs. However, recent 3D contrastive learning approaches either neglect correspondences or fail to maximally capitalize on them. To this end, we propose an extensible 3D contrastive framework (Spade, for Spatial Debiasing) that leverages extracted correspondences to select more effective positive & negative samples for representation learning. Our method learns both globally invariant and locally equivariant representations with downstream segmentation in mind. We also propose separate selection strategies for global & local scopes that tailor to their respective representational requirements. Compared to recent state-of-the-art approaches, Spade shows notable improvements on three downstream segmentation tasks (CT Abdominal Organ, CT Heart, MR Heart).
Unsupervised Feature Clustering Improves Contrastive Representation Learning for Medical Image Segmentation
Nov 15, 2022Abstract:Self-supervised instance discrimination is an effective contrastive pretext task to learn feature representations and address limited medical image annotations. The idea is to make features of transformed versions of the same images similar while forcing all other augmented images' representations to contrast. However, this instance-based contrastive learning leaves performance on the table by failing to maximize feature affinity between images with similar content while counter-productively pushing their representations apart. Recent improvements on this paradigm (e.g., leveraging multi-modal data, different images in longitudinal studies, spatial correspondences) either relied on additional views or made stringent assumptions about data properties, which can sacrifice generalizability and applicability. To address this challenge, we propose a new self-supervised contrastive learning method that uses unsupervised feature clustering to better select positive and negative image samples. More specifically, we produce pseudo-classes by hierarchically clustering features obtained by an auto-encoder in an unsupervised manner, and prevent destructive interference during contrastive learning by avoiding the selection of negatives from the same pseudo-class. Experiments on 2D skin dermoscopic image segmentation and 3D multi-class whole heart CT segmentation demonstrate that our method outperforms state-of-the-art self-supervised contrastive techniques on these tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge