Peixian Liang
VISTA-PATH: An interactive foundation model for pathology image segmentation and quantitative analysis in computational pathology
Jan 23, 2026Abstract:Accurate semantic segmentation for histopathology image is crucial for quantitative tissue analysis and downstream clinical modeling. Recent segmentation foundation models have improved generalization through large-scale pretraining, yet remain poorly aligned with pathology because they treat segmentation as a static visual prediction task. Here we present VISTA-PATH, an interactive, class-aware pathology segmentation foundation model designed to resolve heterogeneous structures, incorporate expert feedback, and produce pixel-level segmentation that are directly meaningful for clinical interpretation. VISTA-PATH jointly conditions segmentation on visual context, semantic tissue descriptions, and optional expert-provided spatial prompts, enabling precise multi-class segmentation across heterogeneous pathology images. To support this paradigm, we curate VISTA-PATH Data, a large-scale pathology segmentation corpus comprising over 1.6 million image-mask-text triplets spanning 9 organs and 93 tissue classes. Across extensive held-out and external benchmarks, VISTA-PATH consistently outperforms existing segmentation foundation models. Importantly, VISTA-PATH supports dynamic human-in-the-loop refinement by propagating sparse, patch-level bounding-box annotation feedback into whole-slide segmentation. Finally, we show that the high-fidelity, class-aware segmentation produced by VISTA-PATH is a preferred model for computational pathology. It improve tissue microenvironment analysis through proposed Tumor Interaction Score (TIS), which exhibits strong and significant associations with patient survival. Together, these results establish VISTA-PATH as a foundation model that elevates pathology image segmentation from a static prediction to an interactive and clinically grounded representation for digital pathology. Source code and demo can be found at https://github.com/zhihuanglab/VISTA-PATH.
SHMC-Net: A Mask-guided Feature Fusion Network for Sperm Head Morphology Classification
Feb 07, 2024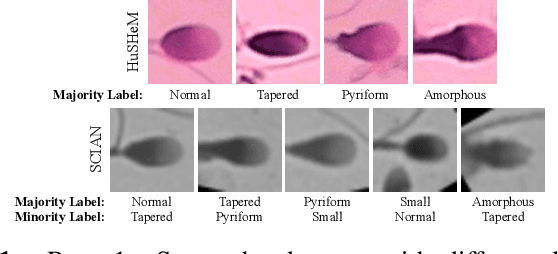
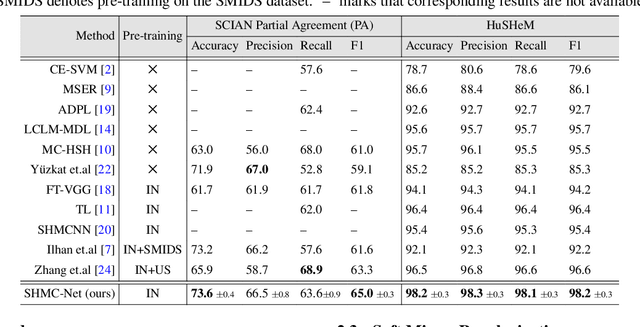
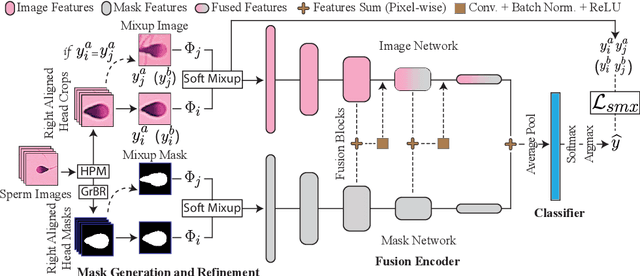
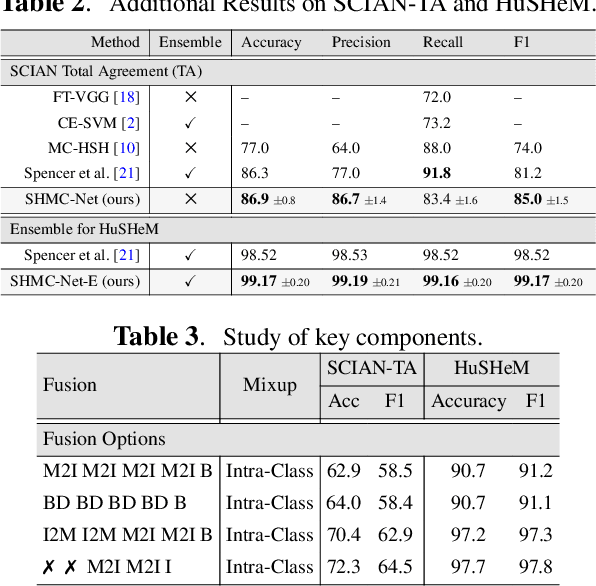
Abstract:Male infertility accounts for about one-third of global infertility cases. Manual assessment of sperm abnormalities through head morphology analysis encounters issues of observer variability and diagnostic discrepancies among experts. Its alternative, Computer-Assisted Semen Analysis (CASA), suffers from low-quality sperm images, small datasets, and noisy class labels. We propose a new approach for sperm head morphology classification, called SHMC-Net, which uses segmentation masks of sperm heads to guide the morphology classification of sperm images. SHMC-Net generates reliable segmentation masks using image priors, refines object boundaries with an efficient graph-based method, and trains an image network with sperm head crops and a mask network with the corresponding masks. In the intermediate stages of the networks, image and mask features are fused with a fusion scheme to better learn morphological features. To handle noisy class labels and regularize training on small datasets, SHMC-Net applies Soft Mixup to combine mixup augmentation and a loss function. We achieve state-of-the-art results on SCIAN and HuSHeM datasets, outperforming methods that use additional pre-training or costly ensembling techniques.
Efficient approximation of Earth Mover's Distance Based on Nearest Neighbor Search
Jan 20, 2024



Abstract:Earth Mover's Distance (EMD) is an important similarity measure between two distributions, used in computer vision and many other application domains. However, its exact calculation is computationally and memory intensive, which hinders its scalability and applicability for large-scale problems. Various approximate EMD algorithms have been proposed to reduce computational costs, but they suffer lower accuracy and may require additional memory usage or manual parameter tuning. In this paper, we present a novel approach, NNS-EMD, to approximate EMD using Nearest Neighbor Search (NNS), in order to achieve high accuracy, low time complexity, and high memory efficiency. The NNS operation reduces the number of data points compared in each NNS iteration and offers opportunities for parallel processing. We further accelerate NNS-EMD via vectorization on GPU, which is especially beneficial for large datasets. We compare NNS-EMD with both the exact EMD and state-of-the-art approximate EMD algorithms on image classification and retrieval tasks. We also apply NNS-EMD to calculate transport mapping and realize color transfer between images. NNS-EMD can be 44x to 135x faster than the exact EMD implementation, and achieves superior accuracy, speedup, and memory efficiency over existing approximate EMD methods.
Input Augmentation with SAM: Boosting Medical Image Segmentation with Segmentation Foundation Model
Apr 22, 2023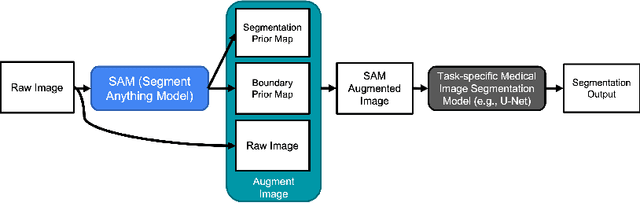
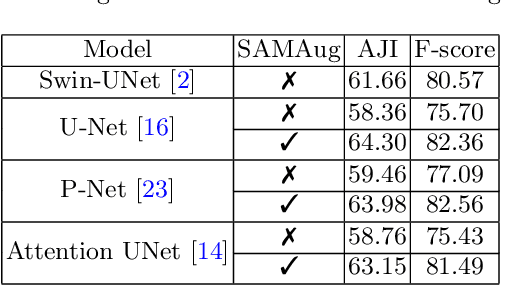
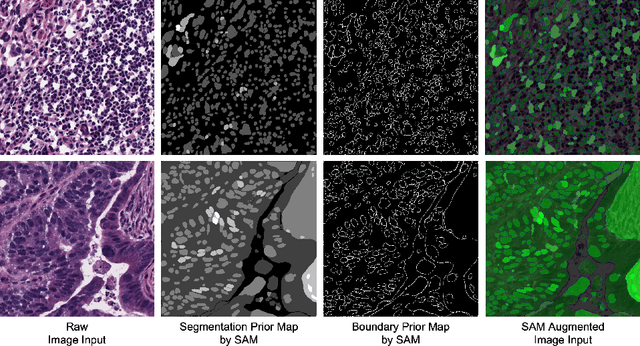
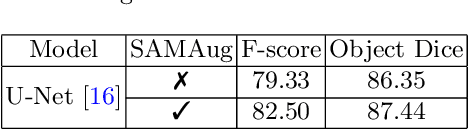
Abstract:The Segment Anything Model (SAM) is a recently developed large model for general-purpose segmentation for computer vision tasks. SAM was trained using 11 million images with over 1 billion masks and can produce segmentation results for a wide range of objects in natural scene images. SAM can be viewed as a general perception model for segmentation (partitioning images into semantically meaningful regions). Thus, how to utilize such a large foundation model for medical image segmentation is an emerging research target. This paper shows that although SAM does not immediately give high-quality segmentation for medical images, its generated masks, features, and stability scores are useful for building and training better medical image segmentation models. In particular, we demonstrate how to use SAM to augment image inputs for a commonly-used medical image segmentation model (e.g., U-Net). Experiments on two datasets show the effectiveness of our proposed method.
A Point in the Right Direction: Vector Prediction for Spatially-aware Self-supervised Volumetric Representation Learning
Nov 15, 2022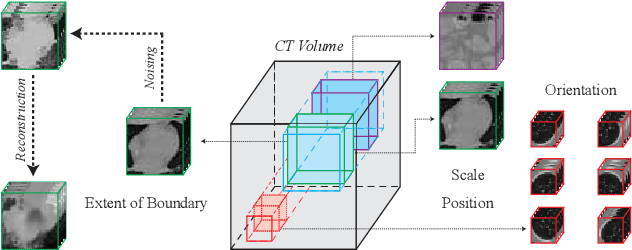
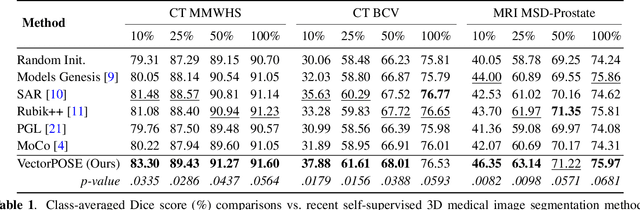
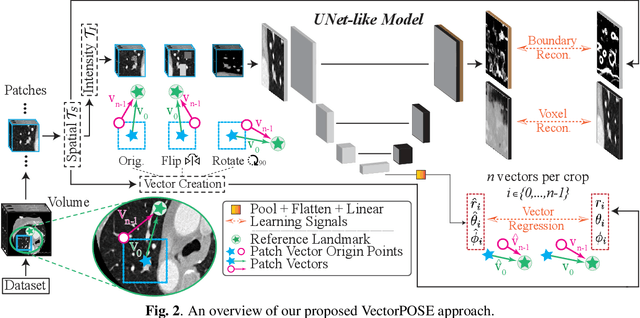
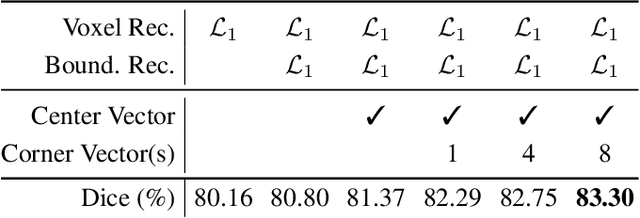
Abstract:High annotation costs and limited labels for dense 3D medical imaging tasks have recently motivated an assortment of 3D self-supervised pretraining methods that improve transfer learning performance. However, these methods commonly lack spatial awareness despite its centrality in enabling effective 3D image analysis. More specifically, position, scale, and orientation are not only informative but also automatically available when generating image crops for training. Yet, to date, no work has proposed a pretext task that distills all key spatial features. To fulfill this need, we develop a new self-supervised method, VectorPOSE, which promotes better spatial understanding with two novel pretext tasks: Vector Prediction (VP) and Boundary-Focused Reconstruction (BFR). VP focuses on global spatial concepts (i.e., properties of 3D patches) while BFR addresses weaknesses of recent reconstruction methods to learn more effective local representations. We evaluate VectorPOSE on three 3D medical image segmentation tasks, showing that it often outperforms state-of-the-art methods, especially in limited annotation settings.
Usable Region Estimate for Assessing Practical Usability of Medical Image Segmentation Models
Jul 01, 2022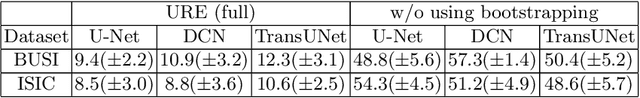
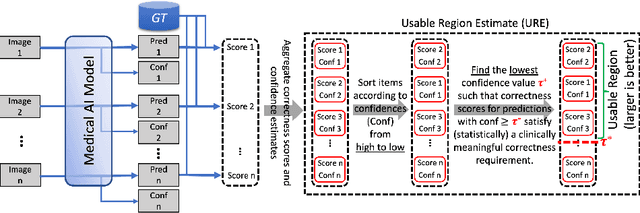
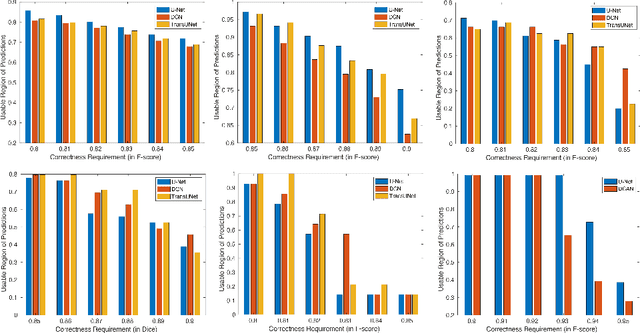
Abstract:We aim to quantitatively measure the practical usability of medical image segmentation models: to what extent, how often, and on which samples a model's predictions can be used/trusted. We first propose a measure, Correctness-Confidence Rank Correlation (CCRC), to capture how predictions' confidence estimates correlate with their correctness scores in rank. A model with a high value of CCRC means its prediction confidences reliably suggest which samples' predictions are more likely to be correct. Since CCRC does not capture the actual prediction correctness, it alone is insufficient to indicate whether a prediction model is both accurate and reliable to use in practice. Therefore, we further propose another method, Usable Region Estimate (URE), which simultaneously quantifies predictions' correctness and reliability of confidence assessments in one estimate. URE provides concrete information on to what extent a model's predictions are usable. In addition, the sizes of usable regions (UR) can be utilized to compare models: A model with a larger UR can be taken as a more usable and hence better model. Experiments on six datasets validate that the proposed evaluation methods perform well, providing a concrete and concise measure for the practical usability of medical image segmentation models. Code is made available at https://github.com/yizhezhang2000/ure.
H-EMD: A Hierarchical Earth Mover's Distance Method for Instance Segmentation
Jun 02, 2022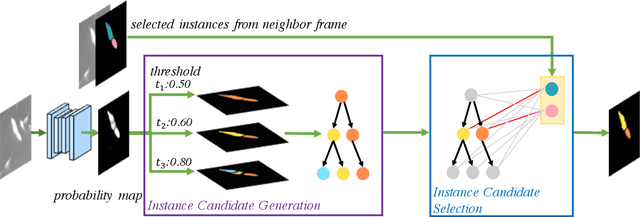
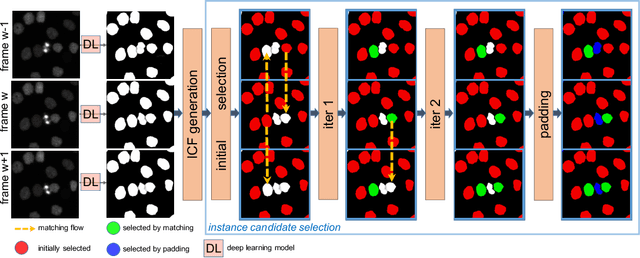
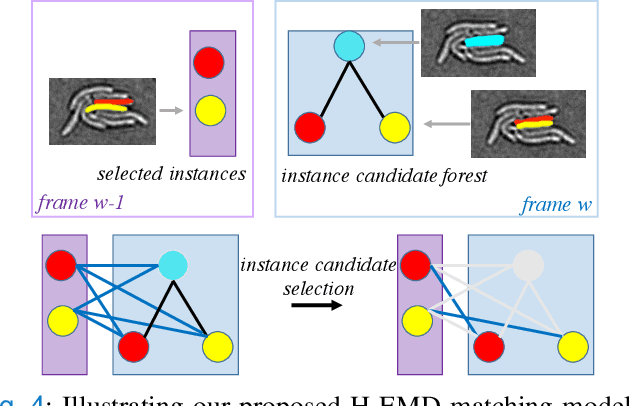

Abstract:Deep learning (DL) based semantic segmentation methods have achieved excellent performance in biomedical image segmentation, producing high quality probability maps to allow extraction of rich instance information to facilitate good instance segmentation. While numerous efforts were put into developing new DL semantic segmentation models, less attention was paid to a key issue of how to effectively explore their probability maps to attain the best possible instance segmentation. We observe that probability maps by DL semantic segmentation models can be used to generate many possible instance candidates, and accurate instance segmentation can be achieved by selecting from them a set of "optimized" candidates as output instances. Further, the generated instance candidates form a well-behaved hierarchical structure (a forest), which allows selecting instances in an optimized manner. Hence, we propose a novel framework, called hierarchical earth mover's distance (H-EMD), for instance segmentation in biomedical 2D+time videos and 3D images, which judiciously incorporates consistent instance selection with semantic-segmentation-generated probability maps. H-EMD contains two main stages. (1) Instance candidate generation: capturing instance-structured information in probability maps by generating many instance candidates in a forest structure. (2) Instance candidate selection: selecting instances from the candidate set for final instance segmentation. We formulate a key instance selection problem on the instance candidate forest as an optimization problem based on the earth mover's distance (EMD), and solve it by integer linear programming. Extensive experiments on eight biomedical video or 3D datasets demonstrate that H-EMD consistently boosts DL semantic segmentation models and is highly competitive with state-of-the-art methods.
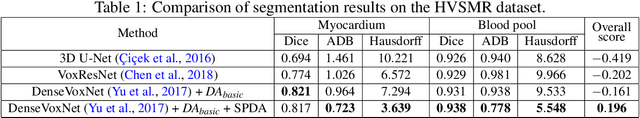
SPDA: Superpixel-based Data Augmentation for Biomedical Image Segmentation
Feb 28, 2019
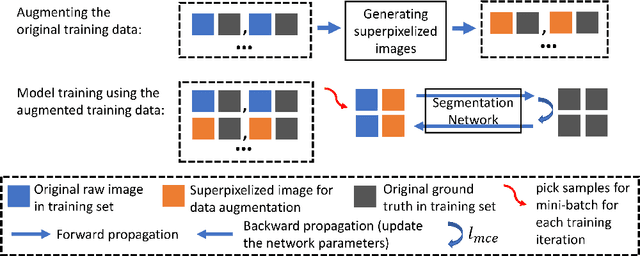
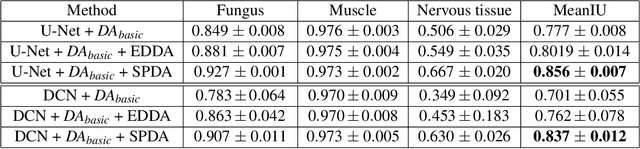
Abstract:Supervised training a deep neural network aims to "teach" the network to mimic human visual perception that is represented by image-and-label pairs in the training data. Superpixelized (SP) images are visually perceivable to humans, but a conventionally trained deep learning model often performs poorly when working on SP images. To better mimic human visual perception, we think it is desirable for the deep learning model to be able to perceive not only raw images but also SP images. In this paper, we propose a new superpixel-based data augmentation (SPDA) method for training deep learning models for biomedical image segmentation. Our method applies a superpixel generation scheme to all the original training images to generate superpixelized images. The SP images thus obtained are then jointly used with the original training images to train a deep learning model. Our experiments of SPDA on four biomedical image datasets show that SPDA is effective and can consistently improve the performance of state-of-the-art fully convolutional networks for biomedical image segmentation in 2D and 3D images. Additional studies also demonstrate that SPDA can practically reduce the generalization gap.
Cascade Decoder: A Universal Decoding Method for Biomedical Image Segmentation
Jan 15, 2019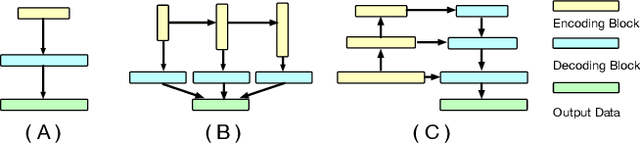
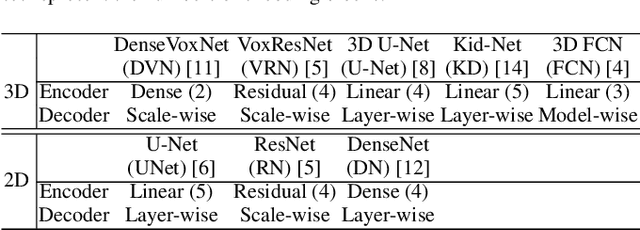
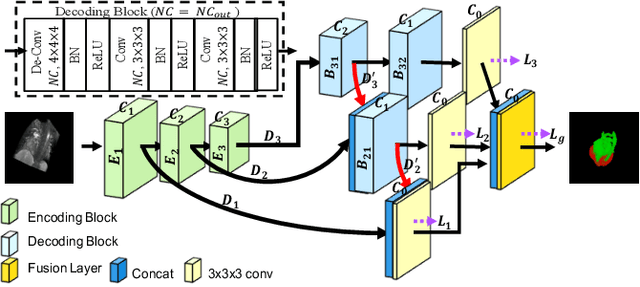
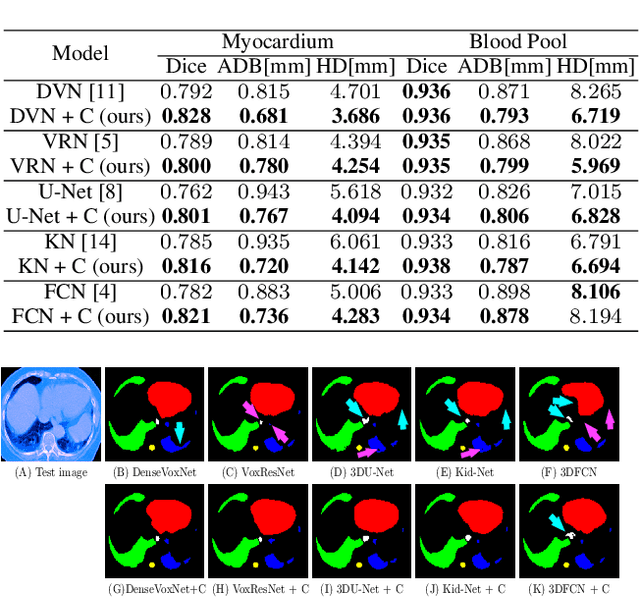
Abstract:The Encoder-Decoder architecture is a main stream deep learning model for biomedical image segmentation. The encoder fully compresses the input and generates encoded features, and the decoder then produces dense predictions using encoded features. However, decoders are still under-explored in such architectures. In this paper, we comprehensively study the state-of-the-art Encoder-Decoder architectures, and propose a new universal decoder, called cascade decoder, to improve semantic segmentation accuracy. Our cascade decoder can be embedded into existing networks and trained altogether in an end-to-end fashion. The cascade decoder structure aims to conduct more effective decoding of hierarchically encoded features and is more compatible with common encoders than the known decoders. We replace the decoders of state-of-the-art models with our cascade decoder for several challenging biomedical image segmentation tasks, and the considerable improvements achieved demonstrate the efficacy of our new decoding method.
CC-Net: Image Complexity Guided Network Compression for Biomedical Image Segmentation
Jan 06, 2019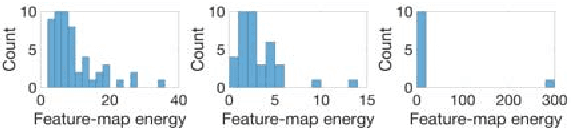
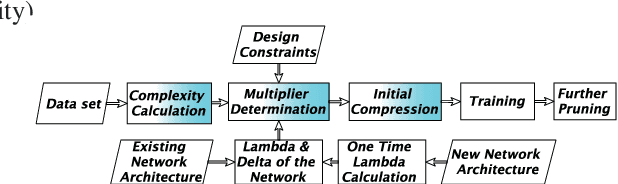
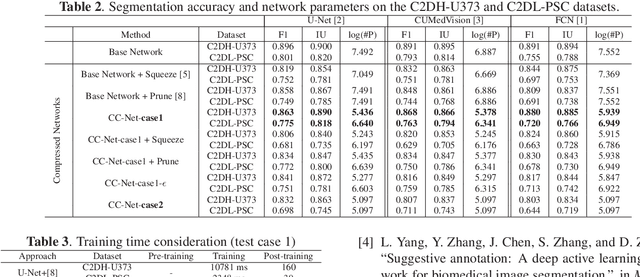
Abstract:Convolutional neural networks (CNNs) for biomedical image analysis are often of very large size, resulting in high memory requirement and high latency of operations. Searching for an acceptable compressed representation of the base CNN for a specific imaging application typically involves a series of time-consuming training/validation experiments to achieve a good compromise between network size and accuracy. To address this challenge, we propose CC-Net, a new image complexity-guided CNN compression scheme for biomedical image segmentation. Given a CNN model, CC-Net predicts the final accuracy of networks of different sizes based on the average image complexity computed from the training data. It then selects a multiplicative factor for producing a desired network with acceptable network accuracy and size. Experiments show that CC-Net is effective for generating compressed segmentation networks, retaining up to 95% of the base network segmentation accuracy and utilizing only 0.1% of trainable parameters of the full-sized networks in the best case.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge