Danny Z Chen
SHMC-Net: A Mask-guided Feature Fusion Network for Sperm Head Morphology Classification
Feb 07, 2024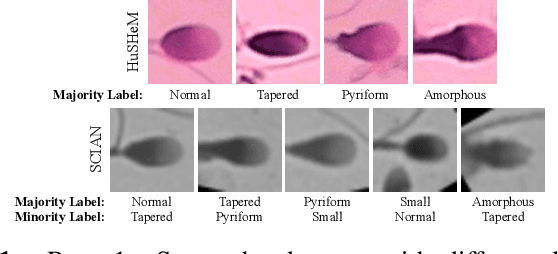
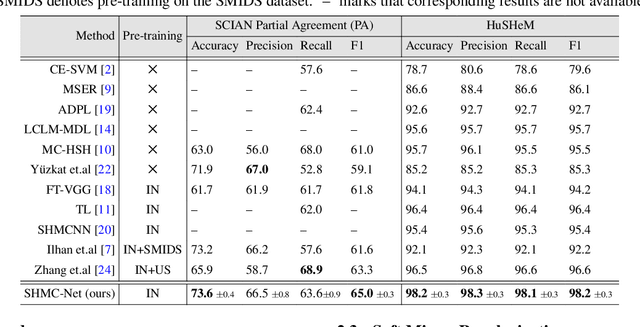
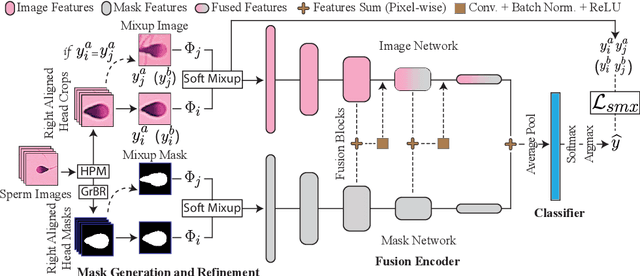
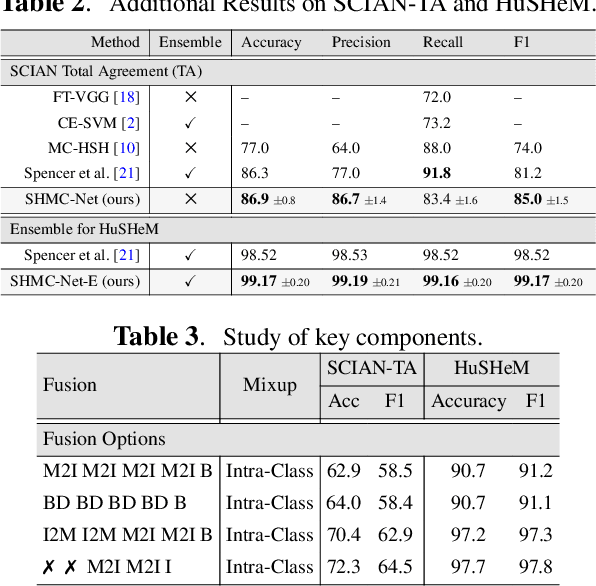
Abstract:Male infertility accounts for about one-third of global infertility cases. Manual assessment of sperm abnormalities through head morphology analysis encounters issues of observer variability and diagnostic discrepancies among experts. Its alternative, Computer-Assisted Semen Analysis (CASA), suffers from low-quality sperm images, small datasets, and noisy class labels. We propose a new approach for sperm head morphology classification, called SHMC-Net, which uses segmentation masks of sperm heads to guide the morphology classification of sperm images. SHMC-Net generates reliable segmentation masks using image priors, refines object boundaries with an efficient graph-based method, and trains an image network with sperm head crops and a mask network with the corresponding masks. In the intermediate stages of the networks, image and mask features are fused with a fusion scheme to better learn morphological features. To handle noisy class labels and regularize training on small datasets, SHMC-Net applies Soft Mixup to combine mixup augmentation and a loss function. We achieve state-of-the-art results on SCIAN and HuSHeM datasets, outperforming methods that use additional pre-training or costly ensembling techniques.
ConUNETR: A Conditional Transformer Network for 3D Micro-CT Embryonic Cartilage Segmentation
Feb 06, 2024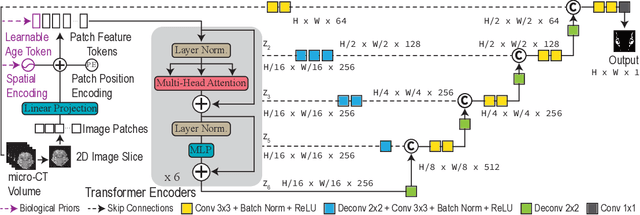
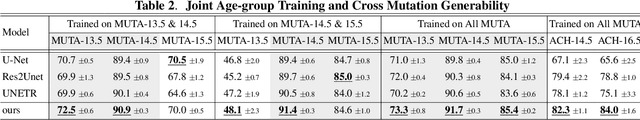
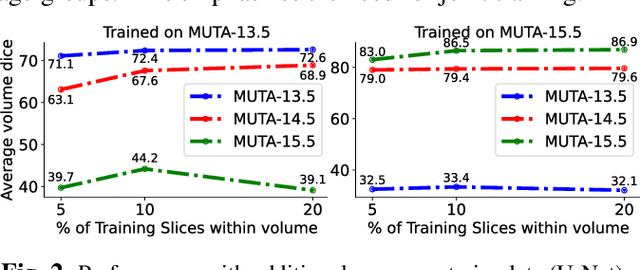
Abstract:Studying the morphological development of cartilaginous and osseous structures is critical to the early detection of life-threatening skeletal dysmorphology. Embryonic cartilage undergoes rapid structural changes within hours, introducing biological variations and morphological shifts that limit the generalization of deep learning-based segmentation models that infer across multiple embryonic age groups. Obtaining individual models for each age group is expensive and less effective, while direct transfer (predicting an age unseen during training) suffers a potential performance drop due to morphological shifts. We propose a novel Transformer-based segmentation model with improved biological priors that better distills morphologically diverse information through conditional mechanisms. This enables a single model to accurately predict cartilage across multiple age groups. Experiments on the mice cartilage dataset show the superiority of our new model compared to other competitive segmentation models. Additional studies on a separate mice cartilage dataset with a distinct mutation show that our model generalizes well and effectively captures age-based cartilage morphology patterns.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge