Suraj Mishra
Bright 4B: Scaling Hyperspherical Learning for Segmentation in 3D Brightfield Microscopy
Dec 27, 2025Abstract:Label-free 3D brightfield microscopy offers a fast and noninvasive way to visualize cellular morphology, yet robust volumetric segmentation still typically depends on fluorescence or heavy post-processing. We address this gap by introducing Bright-4B, a 4 billion parameter foundation model that learns on the unit hypersphere to segment subcellular structures directly from 3D brightfield volumes. Bright-4B combines a hardware-aligned Native Sparse Attention mechanism (capturing local, coarse, and selected global context), depth-width residual HyperConnections that stabilize representation flow, and a soft Mixture-of-Experts for adaptive capacity. A plug-and-play anisotropic patch embed further respects confocal point-spread and axial thinning, enabling geometry-faithful 3D tokenization. The resulting model produces morphology-accurate segmentations of nuclei, mitochondria, and other organelles from brightfield stacks alone--without fluorescence, auxiliary channels, or handcrafted post-processing. Across multiple confocal datasets, Bright-4B preserves fine structural detail across depth and cell types, outperforming contemporary CNN and Transformer baselines. All code, pretrained weights, and models for downstream finetuning will be released to advance large-scale, label-free 3D cell mapping.
Densely Decoded Networks with Adaptive Deep Supervision for Medical Image Segmentation
Feb 05, 2024Abstract:Medical image segmentation using deep neural networks has been highly successful. However, the effectiveness of these networks is often limited by inadequate dense prediction and inability to extract robust features. To achieve refined dense prediction, we propose densely decoded networks (ddn), by selectively introducing 'crutch' network connections. Such 'crutch' connections in each upsampling stage of the network decoder (1) enhance target localization by incorporating high resolution features from the encoder, and (2) improve segmentation by facilitating multi-stage contextual information flow. Further, we present a training strategy based on adaptive deep supervision (ads), which exploits and adapts specific attributes of input dataset, for robust feature extraction. In particular, ads strategically locates and deploys auxiliary supervision, by matching the average input object size with the layer-wise effective receptive fields (lerf) of a network, resulting in a class of ddns. Such inclusion of 'companion objective' from a specific hidden layer, helps the model pay close attention to some distinct input-dependent features, which the network might otherwise 'ignore' during training. Our new networks and training strategy are validated on 4 diverse datasets of different modalities, demonstrating their effectiveness.
Data-Driven Deep Supervision for Skin Lesion Classification
Sep 04, 2022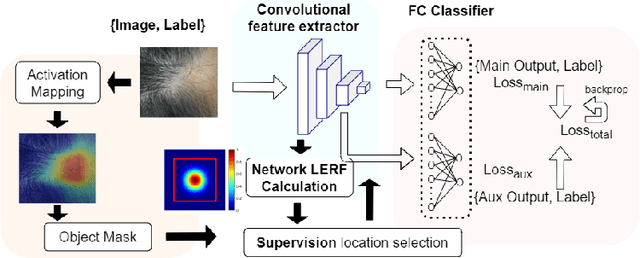
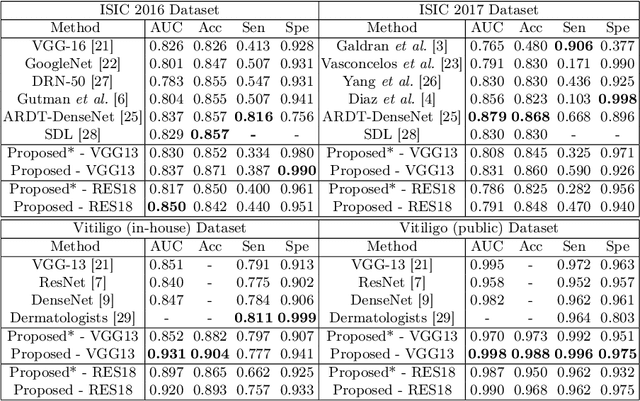
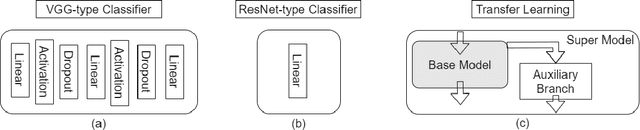
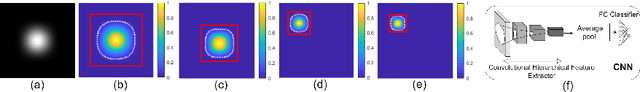
Abstract:Automatic classification of pigmented, non-pigmented, and depigmented non-melanocytic skin lesions have garnered lots of attention in recent years. However, imaging variations in skin texture, lesion shape, depigmentation contrast, lighting condition, etc. hinder robust feature extraction, affecting classification accuracy. In this paper, we propose a new deep neural network that exploits input data for robust feature extraction. Specifically, we analyze the convolutional network's behavior (field-of-view) to find the location of deep supervision for improved feature extraction. To achieve this, first, we perform activation mapping to generate an object mask, highlighting the input regions most critical for classification output generation. Then the network layer whose layer-wise effective receptive field matches the approximated object shape in the object mask is selected as our focus for deep supervision. Utilizing different types of convolutional feature extractors and classifiers on three melanoma detection datasets and two vitiligo detection datasets, we verify the effectiveness of our new method.
Usable Region Estimate for Assessing Practical Usability of Medical Image Segmentation Models
Jul 01, 2022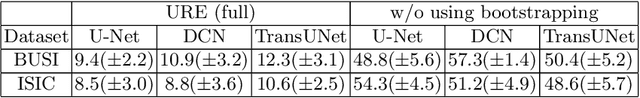
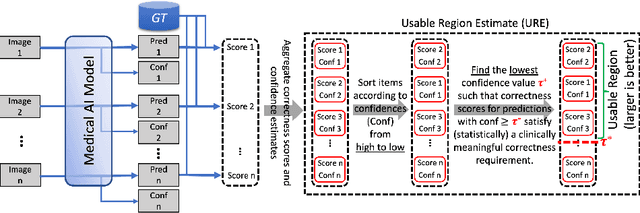
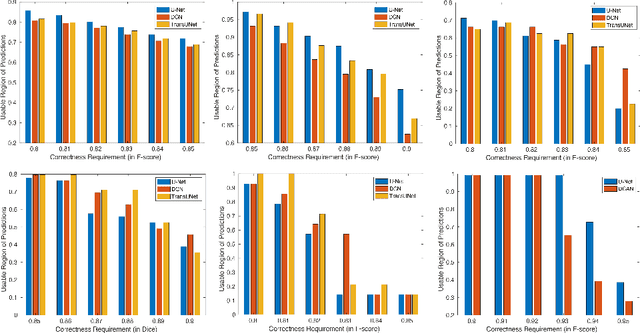
Abstract:We aim to quantitatively measure the practical usability of medical image segmentation models: to what extent, how often, and on which samples a model's predictions can be used/trusted. We first propose a measure, Correctness-Confidence Rank Correlation (CCRC), to capture how predictions' confidence estimates correlate with their correctness scores in rank. A model with a high value of CCRC means its prediction confidences reliably suggest which samples' predictions are more likely to be correct. Since CCRC does not capture the actual prediction correctness, it alone is insufficient to indicate whether a prediction model is both accurate and reliable to use in practice. Therefore, we further propose another method, Usable Region Estimate (URE), which simultaneously quantifies predictions' correctness and reliability of confidence assessments in one estimate. URE provides concrete information on to what extent a model's predictions are usable. In addition, the sizes of usable regions (UR) can be utilized to compare models: A model with a larger UR can be taken as a more usable and hence better model. Experiments on six datasets validate that the proposed evaluation methods perform well, providing a concrete and concise measure for the practical usability of medical image segmentation models. Code is made available at https://github.com/yizhezhang2000/ure.
Image Complexity Guided Network Compression for Biomedical Image Segmentation
Jul 06, 2021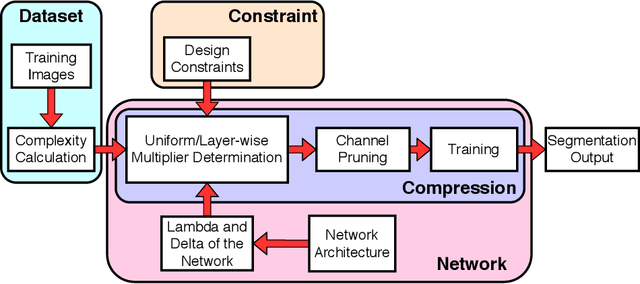
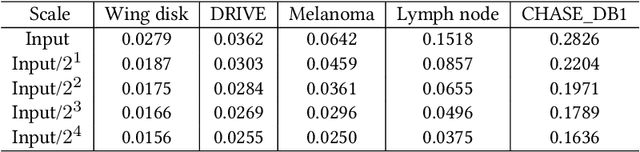
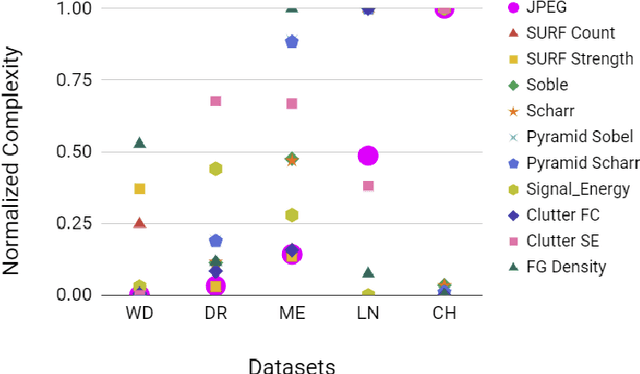
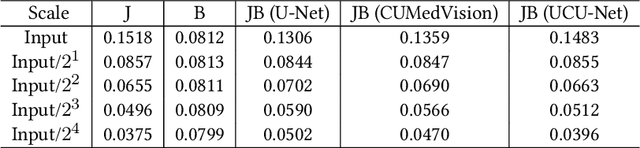
Abstract:Compression is a standard procedure for making convolutional neural networks (CNNs) adhere to some specific computing resource constraints. However, searching for a compressed architecture typically involves a series of time-consuming training/validation experiments to determine a good compromise between network size and performance accuracy. To address this, we propose an image complexity-guided network compression technique for biomedical image segmentation. Given any resource constraints, our framework utilizes data complexity and network architecture to quickly estimate a compressed model which does not require network training. Specifically, we map the dataset complexity to the target network accuracy degradation caused by compression. Such mapping enables us to predict the final accuracy for different network sizes, based on the computed dataset complexity. Thus, one may choose a solution that meets both the network size and segmentation accuracy requirements. Finally, the mapping is used to determine the convolutional layer-wise multiplicative factor for generating a compressed network. We conduct experiments using 5 datasets, employing 3 commonly-used CNN architectures for biomedical image segmentation as representative networks. Our proposed framework is shown to be effective for generating compressed segmentation networks, retaining up to $\approx 95\%$ of the full-sized network segmentation accuracy, and at the same time, utilizing $\approx 32x$ fewer network trainable weights (average reduction) of the full-sized networks.
Objective-Dependent Uncertainty Driven Retinal Vessel Segmentation
Apr 17, 2021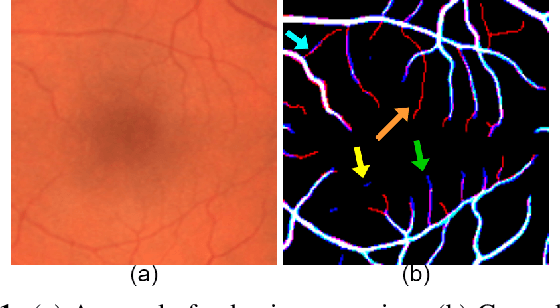
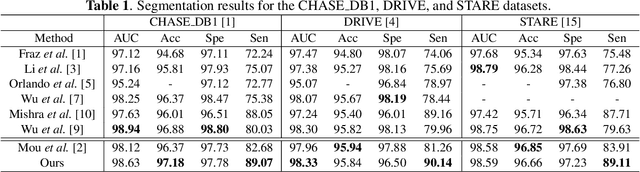
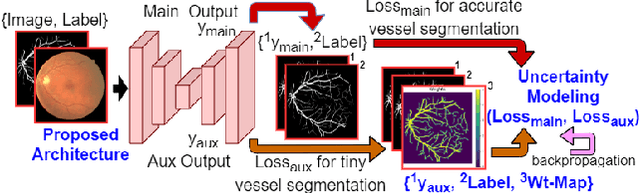
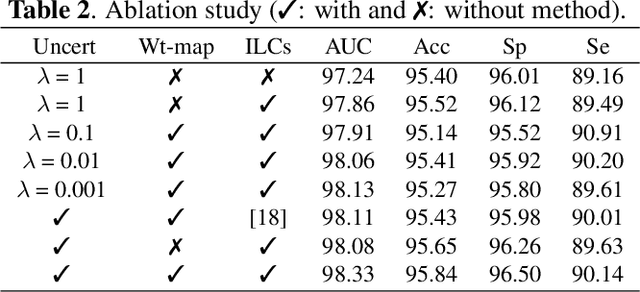
Abstract:From diagnosing neovascular diseases to detecting white matter lesions, accurate tiny vessel segmentation in fundus images is critical. Promising results for accurate vessel segmentation have been known. However, their effectiveness in segmenting tiny vessels is still limited. In this paper, we study retinal vessel segmentation by incorporating tiny vessel segmentation into our framework for the overall accurate vessel segmentation. To achieve this, we propose a new deep convolutional neural network (CNN) which divides vessel segmentation into two separate objectives. Specifically, we consider the overall accurate vessel segmentation and tiny vessel segmentation as two individual objectives. Then, by exploiting the objective-dependent (homoscedastic) uncertainty, we enable the network to learn both objectives simultaneously. Further, to improve the individual objectives, we propose: (a) a vessel weight map based auxiliary loss for enhancing tiny vessel connectivity (i.e., improving tiny vessel segmentation), and (b) an enhanced encoder-decoder architecture for improved localization (i.e., for accurate vessel segmentation). Using 3 public retinal vessel segmentation datasets (CHASE_DB1, DRIVE, and STARE), we verify the superiority of our proposed framework in segmenting tiny vessels (8.3% average improvement in sensitivity) while achieving better area under the receiver operating characteristic curve (AUC) compared to state-of-the-art methods.
CC-Net: Image Complexity Guided Network Compression for Biomedical Image Segmentation
Jan 06, 2019
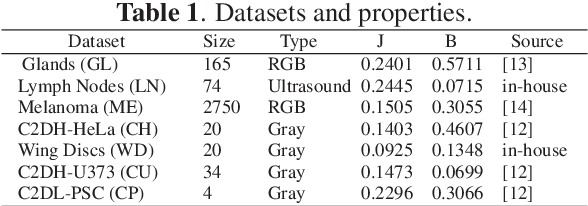
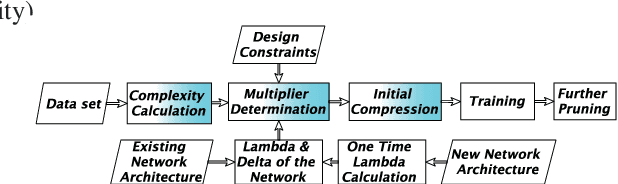
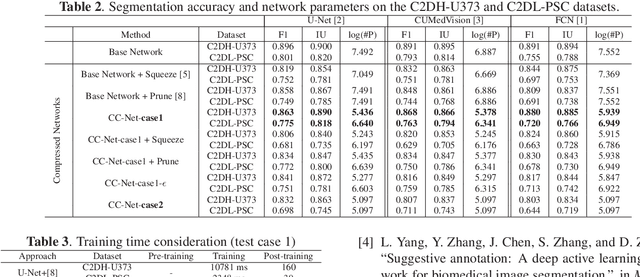
Abstract:Convolutional neural networks (CNNs) for biomedical image analysis are often of very large size, resulting in high memory requirement and high latency of operations. Searching for an acceptable compressed representation of the base CNN for a specific imaging application typically involves a series of time-consuming training/validation experiments to achieve a good compromise between network size and accuracy. To address this challenge, we propose CC-Net, a new image complexity-guided CNN compression scheme for biomedical image segmentation. Given a CNN model, CC-Net predicts the final accuracy of networks of different sizes based on the average image complexity computed from the training data. It then selects a multiplicative factor for producing a desired network with acceptable network accuracy and size. Experiments show that CC-Net is effective for generating compressed segmentation networks, retaining up to 95% of the base network segmentation accuracy and utilizing only 0.1% of trainable parameters of the full-sized networks in the best case.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge