Jiahuan Yan
Spatial RoboGrasp: Generalized Robotic Grasping Control Policy
May 27, 2025



Abstract:Achieving generalizable and precise robotic manipulation across diverse environments remains a critical challenge, largely due to limitations in spatial perception. While prior imitation-learning approaches have made progress, their reliance on raw RGB inputs and handcrafted features often leads to overfitting and poor 3D reasoning under varied lighting, occlusion, and object conditions. In this paper, we propose a unified framework that couples robust multimodal perception with reliable grasp prediction. Our architecture fuses domain-randomized augmentation, monocular depth estimation, and a depth-aware 6-DoF Grasp Prompt into a single spatial representation for downstream action planning. Conditioned on this encoding and a high-level task prompt, our diffusion-based policy yields precise action sequences, achieving up to 40% improvement in grasp success and 45% higher task success rates under environmental variation. These results demonstrate that spatially grounded perception, paired with diffusion-based imitation learning, offers a scalable and robust solution for general-purpose robotic grasping.
CoinRobot: Generalized End-to-end Robotic Learning for Physical Intelligence
Mar 07, 2025Abstract:Physical intelligence holds immense promise for advancing embodied intelligence, enabling robots to acquire complex behaviors from demonstrations. However, achieving generalization and transfer across diverse robotic platforms and environments requires careful design of model architectures, training strategies, and data diversity. Meanwhile existing systems often struggle with scalability, adaptability to heterogeneous hardware, and objective evaluation in real-world settings. We present a generalized end-to-end robotic learning framework designed to bridge this gap. Our framework introduces a unified architecture that supports cross-platform adaptability, enabling seamless deployment across industrial-grade robots, collaborative arms, and novel embodiments without task-specific modifications. By integrating multi-task learning with streamlined network designs, it achieves more robust performance than conventional approaches, while maintaining compatibility with varying sensor configurations and action spaces. We validate our framework through extensive experiments on seven manipulation tasks. Notably, Diffusion-based models trained in our framework demonstrated superior performance and generalizability compared to the LeRobot framework, achieving performance improvements across diverse robotic platforms and environmental conditions.
RoboGrasp: A Universal Grasping Policy for Robust Robotic Control
Feb 05, 2025



Abstract:Imitation learning and world models have shown significant promise in advancing generalizable robotic learning, with robotic grasping remaining a critical challenge for achieving precise manipulation. Existing methods often rely heavily on robot arm state data and RGB images, leading to overfitting to specific object shapes or positions. To address these limitations, we propose RoboGrasp, a universal grasping policy framework that integrates pretrained grasp detection models with robotic learning. By leveraging robust visual guidance from object detection and segmentation tasks, RoboGrasp significantly enhances grasp precision, stability, and generalizability, achieving up to 34% higher success rates in few-shot learning and grasping box prompt tasks. Built on diffusion-based methods, RoboGrasp is adaptable to various robotic learning paradigms, enabling precise and reliable manipulation across diverse and complex scenarios. This framework represents a scalable and versatile solution for tackling real-world challenges in robotic grasping.
Spatially Visual Perception for End-to-End Robotic Learning
Nov 26, 2024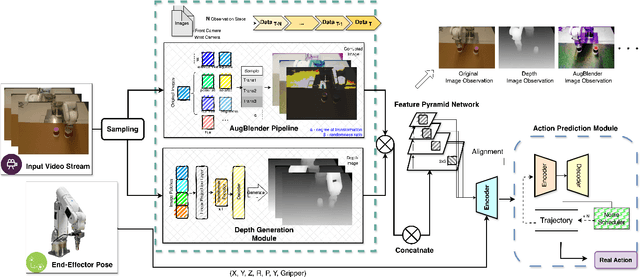
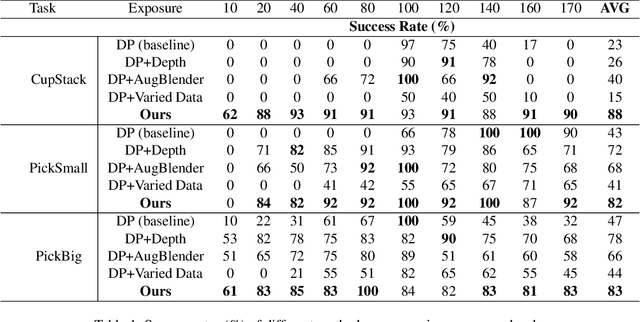
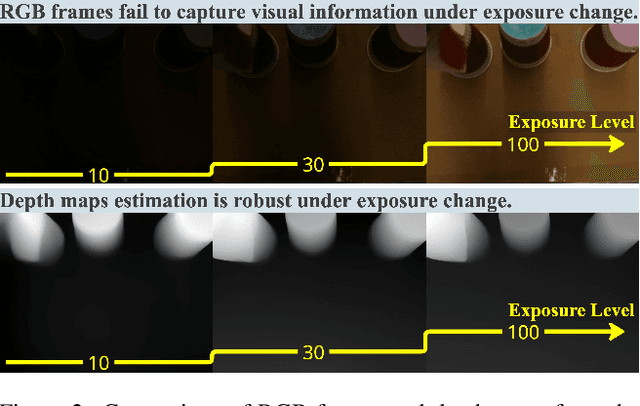
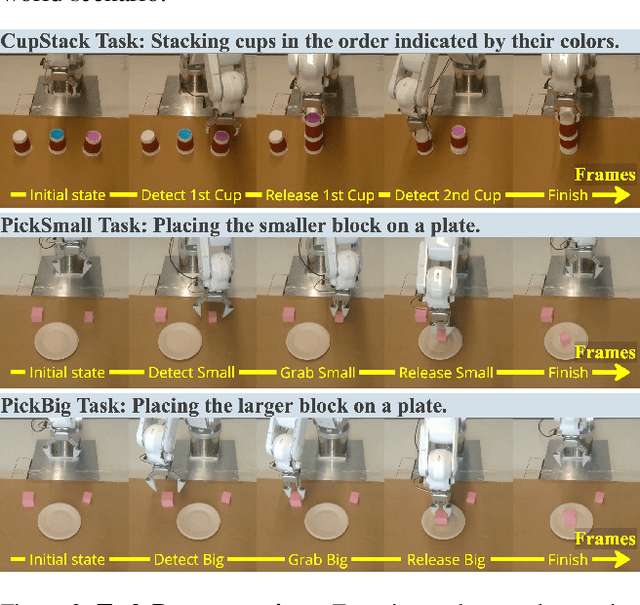
Abstract:Recent advances in imitation learning have shown significant promise for robotic control and embodied intelligence. However, achieving robust generalization across diverse mounted camera observations remains a critical challenge. In this paper, we introduce a video-based spatial perception framework that leverages 3D spatial representations to address environmental variability, with a focus on handling lighting changes. Our approach integrates a novel image augmentation technique, AugBlender, with a state-of-the-art monocular depth estimation model trained on internet-scale data. Together, these components form a cohesive system designed to enhance robustness and adaptability in dynamic scenarios. Our results demonstrate that our approach significantly boosts the success rate across diverse camera exposures, where previous models experience performance collapse. Our findings highlight the potential of video-based spatial perception models in advancing robustness for end-to-end robotic learning, paving the way for scalable, low-cost solutions in embodied intelligence.
Bridge-IF: Learning Inverse Protein Folding with Markov Bridges
Nov 04, 2024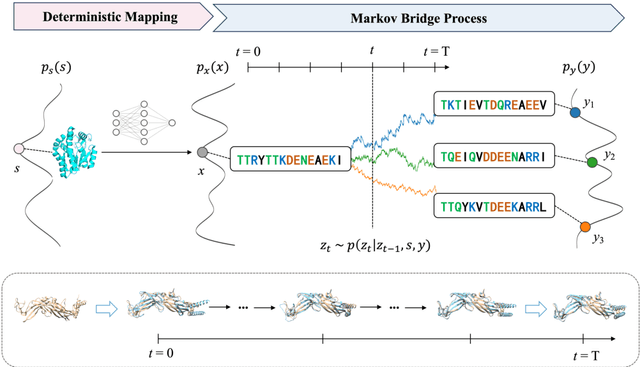
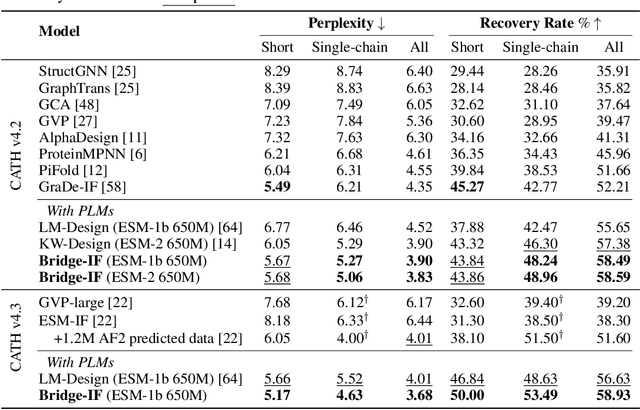
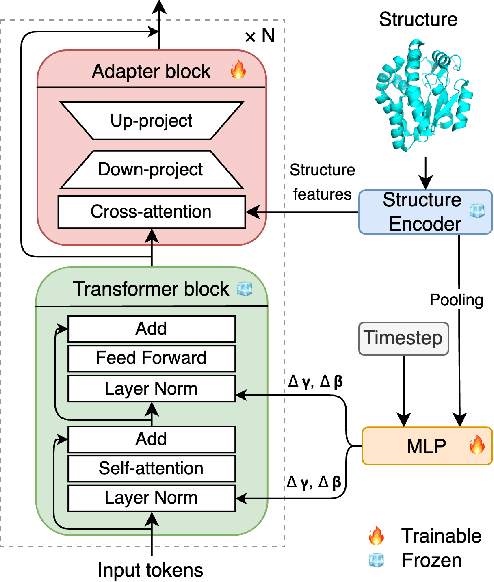
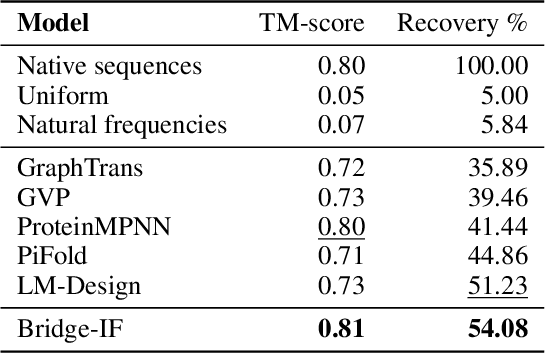
Abstract:Inverse protein folding is a fundamental task in computational protein design, which aims to design protein sequences that fold into the desired backbone structures. While the development of machine learning algorithms for this task has seen significant success, the prevailing approaches, which predominantly employ a discriminative formulation, frequently encounter the error accumulation issue and often fail to capture the extensive variety of plausible sequences. To fill these gaps, we propose Bridge-IF, a generative diffusion bridge model for inverse folding, which is designed to learn the probabilistic dependency between the distributions of backbone structures and protein sequences. Specifically, we harness an expressive structure encoder to propose a discrete, informative prior derived from structures, and establish a Markov bridge to connect this prior with native sequences. During the inference stage, Bridge-IF progressively refines the prior sequence, culminating in a more plausible design. Moreover, we introduce a reparameterization perspective on Markov bridge models, from which we derive a simplified loss function that facilitates more effective training. We also modulate protein language models (PLMs) with structural conditions to precisely approximate the Markov bridge process, thereby significantly enhancing generation performance while maintaining parameter-efficient training. Extensive experiments on well-established benchmarks demonstrate that Bridge-IF predominantly surpasses existing baselines in sequence recovery and excels in the design of plausible proteins with high foldability. The code is available at https://github.com/violet-sto/Bridge-IF.
Generalized Robot Learning Framework
Sep 18, 2024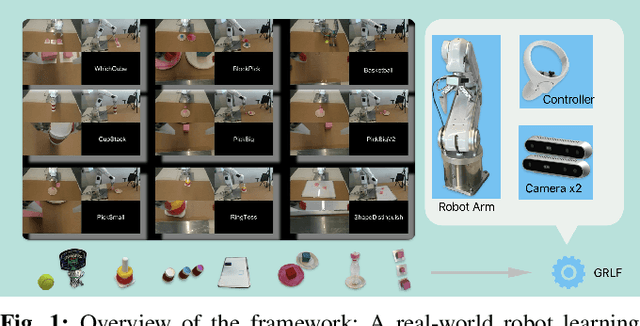
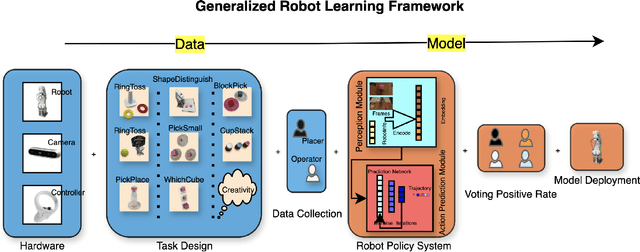
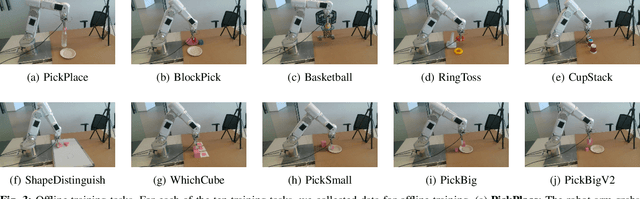
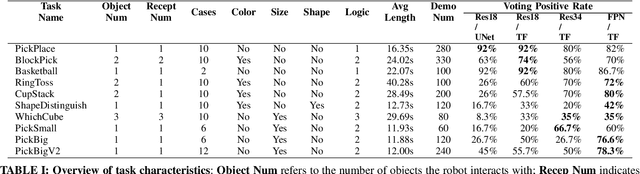
Abstract:Imitation based robot learning has recently gained significant attention in the robotics field due to its theoretical potential for transferability and generalizability. However, it remains notoriously costly, both in terms of hardware and data collection, and deploying it in real-world environments demands meticulous setup of robots and precise experimental conditions. In this paper, we present a low-cost robot learning framework that is both easily reproducible and transferable to various robots and environments. We demonstrate that deployable imitation learning can be successfully applied even to industrial-grade robots, not just expensive collaborative robotic arms. Furthermore, our results show that multi-task robot learning is achievable with simple network architectures and fewer demonstrations than previously thought necessary. As the current evaluating method is almost subjective when it comes to real-world manipulation tasks, we propose Voting Positive Rate (VPR) - a novel evaluation strategy that provides a more objective assessment of performance. We conduct an extensive comparison of success rates across various self-designed tasks to validate our approach. To foster collaboration and support the robot learning community, we have open-sourced all relevant datasets and model checkpoints, available at huggingface.co/ZhiChengAI.
Team up GBDTs and DNNs: Advancing Efficient and Effective Tabular Prediction with Tree-hybrid MLPs
Jul 13, 2024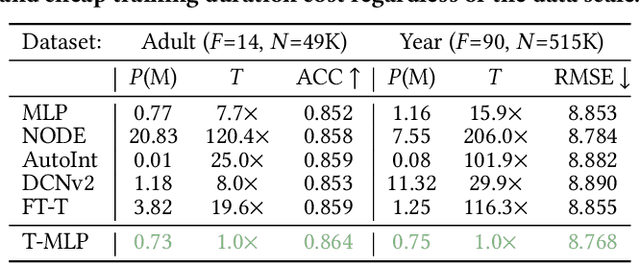
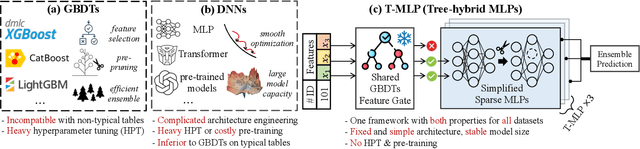
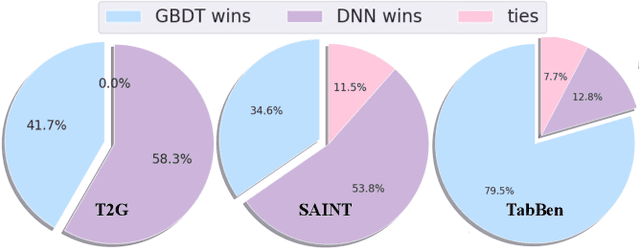
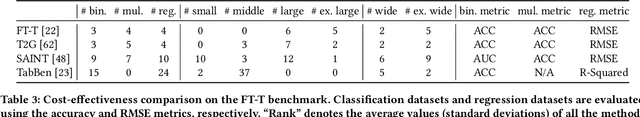
Abstract:Tabular datasets play a crucial role in various applications. Thus, developing efficient, effective, and widely compatible prediction algorithms for tabular data is important. Currently, two prominent model types, Gradient Boosted Decision Trees (GBDTs) and Deep Neural Networks (DNNs), have demonstrated performance advantages on distinct tabular prediction tasks. However, selecting an effective model for a specific tabular dataset is challenging, often demanding time-consuming hyperparameter tuning. To address this model selection dilemma, this paper proposes a new framework that amalgamates the advantages of both GBDTs and DNNs, resulting in a DNN algorithm that is as efficient as GBDTs and is competitively effective regardless of dataset preferences for GBDTs or DNNs. Our idea is rooted in an observation that deep learning (DL) offers a larger parameter space that can represent a well-performing GBDT model, yet the current back-propagation optimizer struggles to efficiently discover such optimal functionality. On the other hand, during GBDT development, hard tree pruning, entropy-driven feature gate, and model ensemble have proved to be more adaptable to tabular data. By combining these key components, we present a Tree-hybrid simple MLP (T-MLP). In our framework, a tensorized, rapidly trained GBDT feature gate, a DNN architecture pruning approach, as well as a vanilla back-propagation optimizer collaboratively train a randomly initialized MLP model. Comprehensive experiments show that T-MLP is competitive with extensively tuned DNNs and GBDTs in their dominating tabular benchmarks (88 datasets) respectively, all achieved with compact model storage and significantly reduced training duration.
Personalized Heart Disease Detection via ECG Digital Twin Generation
Apr 17, 2024Abstract:Heart diseases rank among the leading causes of global mortality, demonstrating a crucial need for early diagnosis and intervention. Most traditional electrocardiogram (ECG) based automated diagnosis methods are trained at population level, neglecting the customization of personalized ECGs to enhance individual healthcare management. A potential solution to address this limitation is to employ digital twins to simulate symptoms of diseases in real patients. In this paper, we present an innovative prospective learning approach for personalized heart disease detection, which generates digital twins of healthy individuals' anomalous ECGs and enhances the model sensitivity to the personalized symptoms. In our approach, a vector quantized feature separator is proposed to locate and isolate the disease symptom and normal segments in ECG signals with ECG report guidance. Thus, the ECG digital twins can simulate specific heart diseases used to train a personalized heart disease detection model. Experiments demonstrate that our approach not only excels in generating high-fidelity ECG signals but also improves personalized heart disease detection. Moreover, our approach ensures robust privacy protection, safeguarding patient data in model development.
Making Pre-trained Language Models Great on Tabular Prediction
Mar 12, 2024



Abstract:The transferability of deep neural networks (DNNs) has made significant progress in image and language processing. However, due to the heterogeneity among tables, such DNN bonus is still far from being well exploited on tabular data prediction (e.g., regression or classification tasks). Condensing knowledge from diverse domains, language models (LMs) possess the capability to comprehend feature names from various tables, potentially serving as versatile learners in transferring knowledge across distinct tables and diverse prediction tasks, but their discrete text representation space is inherently incompatible with numerical feature values in tables. In this paper, we present TP-BERTa, a specifically pre-trained LM for tabular data prediction. Concretely, a novel relative magnitude tokenization converts scalar numerical feature values to finely discrete, high-dimensional tokens, and an intra-feature attention approach integrates feature values with the corresponding feature names. Comprehensive experiments demonstrate that our pre-trained TP-BERTa leads the performance among tabular DNNs and is competitive with Gradient Boosted Decision Tree models in typical tabular data regime.
SERVAL: Synergy Learning between Vertical Models and LLMs towards Oracle-Level Zero-shot Medical Prediction
Mar 03, 2024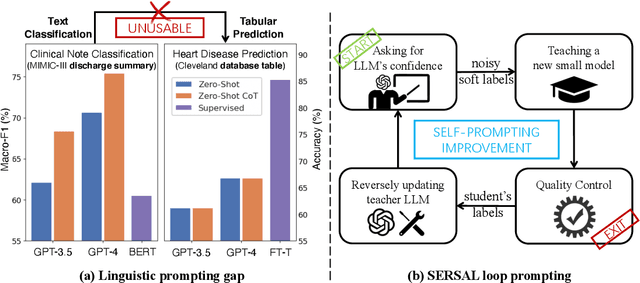

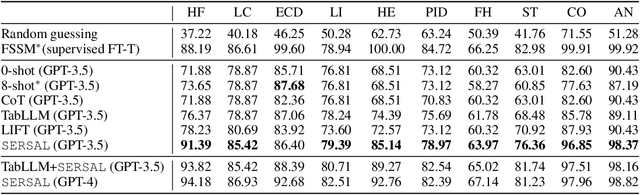
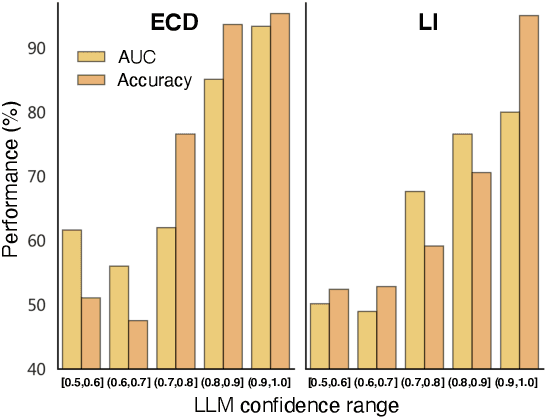
Abstract:Recent development of large language models (LLMs) has exhibited impressive zero-shot proficiency on generic and common sense questions. However, LLMs' application on domain-specific vertical questions still lags behind, primarily due to the humiliation problems and deficiencies in vertical knowledge. Furthermore, the vertical data annotation process often requires labor-intensive expert involvement, thereby presenting an additional challenge in enhancing the model's vertical capabilities. In this paper, we propose SERVAL, a synergy learning pipeline designed for unsupervised development of vertical capabilities in both LLMs and small models by mutual enhancement. Specifically, SERVAL utilizes the LLM's zero-shot outputs as annotations, leveraging its confidence to teach a robust vertical model from scratch. Reversely, the trained vertical model guides the LLM fine-tuning to enhance its zero-shot capability, progressively improving both models through an iterative process. In medical domain, known for complex vertical knowledge and costly annotations, comprehensive experiments show that, without access to any gold labels, SERVAL with the synergy learning of OpenAI GPT-3.5 and a simple model attains fully-supervised competitive performance across ten widely used medical datasets. These datasets represent vertically specialized medical diagnostic scenarios (e.g., diabetes, heart diseases, COVID-19), highlighting the potential of SERVAL in refining the vertical capabilities of LLMs and training vertical models from scratch, all achieved without the need for annotations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge