Jialu Wu
ProtFlow: Fast Protein Sequence Design via Flow Matching on Compressed Protein Language Model Embeddings
Apr 15, 2025Abstract:The design of protein sequences with desired functionalities is a fundamental task in protein engineering. Deep generative methods, such as autoregressive models and diffusion models, have greatly accelerated the discovery of novel protein sequences. However, these methods mainly focus on local or shallow residual semantics and suffer from low inference efficiency, large modeling space and high training cost. To address these challenges, we introduce ProtFlow, a fast flow matching-based protein sequence design framework that operates on embeddings derived from semantically meaningful latent space of protein language models. By compressing and smoothing the latent space, ProtFlow enhances performance while training on limited computational resources. Leveraging reflow techniques, ProtFlow enables high-quality single-step sequence generation. Additionally, we develop a joint design pipeline for the design scene of multichain proteins. We evaluate ProtFlow across diverse protein design tasks, including general peptides and long-chain proteins, antimicrobial peptides, and antibodies. Experimental results demonstrate that ProtFlow outperforms task-specific methods in these applications, underscoring its potential and broad applicability in computational protein sequence design and analysis.
S$^2$ALM: Sequence-Structure Pre-trained Large Language Model for Comprehensive Antibody Representation Learning
Nov 20, 2024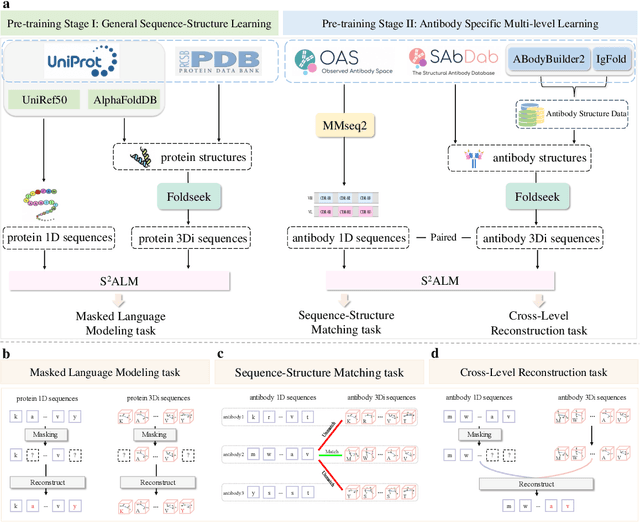
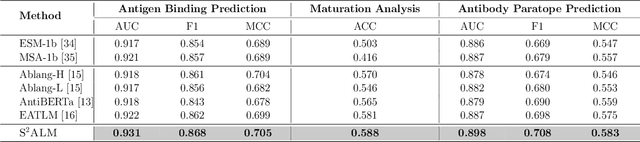
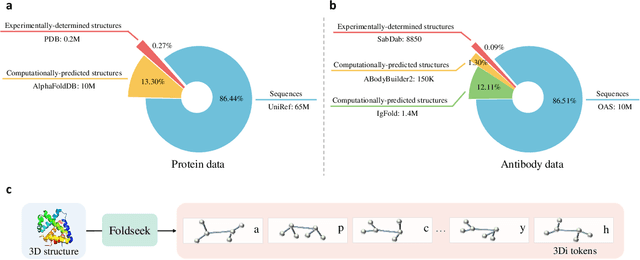
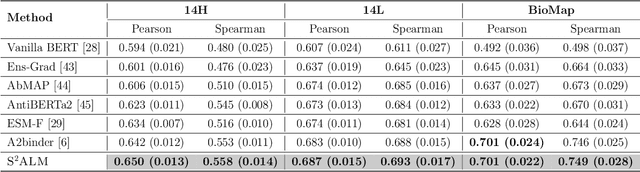
Abstract:Antibodies safeguard our health through their precise and potent binding to specific antigens, demonstrating promising therapeutic efficacy in the treatment of numerous diseases, including COVID-19. Recent advancements in biomedical language models have shown the great potential to interpret complex biological structures and functions. However, existing antibody specific models have a notable limitation that they lack explicit consideration for antibody structural information, despite the fact that both 1D sequence and 3D structure carry unique and complementary insights into antibody behavior and functionality. This paper proposes Sequence-Structure multi-level pre-trained Antibody Language Model (S$^2$ALM), combining holistic sequential and structural information in one unified, generic antibody foundation model. We construct a hierarchical pre-training paradigm incorporated with two customized multi-level training objectives to facilitate the modeling of comprehensive antibody representations. S$^2$ALM's representation space uncovers inherent functional binding mechanisms, biological evolution properties and structural interaction patterns. Pre-trained over 75 million sequences and 11.7 million structures, S$^2$ALM can be adopted for diverse downstream tasks: accurately predicting antigen-antibody binding affinities, precisely distinguishing B cell maturation stages, identifying antibody crucial binding positions, and specifically designing novel coronavirus-binding antibodies. Remarkably, S$^2$ALM outperforms well-established and renowned baselines and sets new state-of-the-art performance across extensive antibody specific understanding and generation tasks. S$^2$ALM's ability to model comprehensive and generalized representations further positions its potential to advance real-world therapeutic antibody development, potentially addressing unmet academic, industrial, and clinical needs.
Bridge-IF: Learning Inverse Protein Folding with Markov Bridges
Nov 04, 2024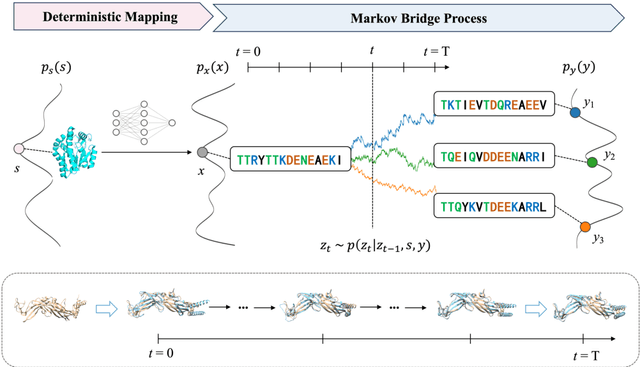
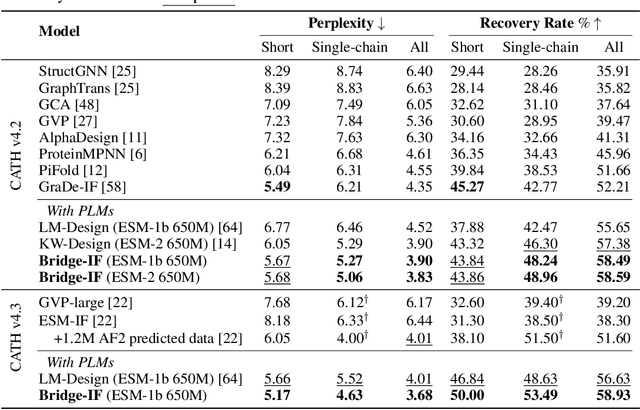
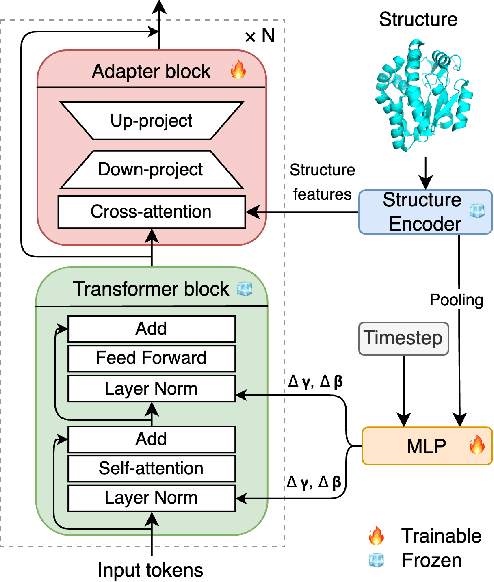
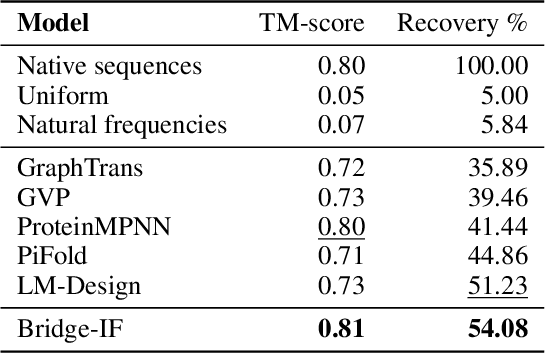
Abstract:Inverse protein folding is a fundamental task in computational protein design, which aims to design protein sequences that fold into the desired backbone structures. While the development of machine learning algorithms for this task has seen significant success, the prevailing approaches, which predominantly employ a discriminative formulation, frequently encounter the error accumulation issue and often fail to capture the extensive variety of plausible sequences. To fill these gaps, we propose Bridge-IF, a generative diffusion bridge model for inverse folding, which is designed to learn the probabilistic dependency between the distributions of backbone structures and protein sequences. Specifically, we harness an expressive structure encoder to propose a discrete, informative prior derived from structures, and establish a Markov bridge to connect this prior with native sequences. During the inference stage, Bridge-IF progressively refines the prior sequence, culminating in a more plausible design. Moreover, we introduce a reparameterization perspective on Markov bridge models, from which we derive a simplified loss function that facilitates more effective training. We also modulate protein language models (PLMs) with structural conditions to precisely approximate the Markov bridge process, thereby significantly enhancing generation performance while maintaining parameter-efficient training. Extensive experiments on well-established benchmarks demonstrate that Bridge-IF predominantly surpasses existing baselines in sequence recovery and excels in the design of plausible proteins with high foldability. The code is available at https://github.com/violet-sto/Bridge-IF.
Multi-Modal CLIP-Informed Protein Editing
Jul 27, 2024Abstract:Proteins govern most biological functions essential for life, but achieving controllable protein discovery and optimization remains challenging. Recently, machine learning-assisted protein editing (MLPE) has shown promise in accelerating optimization cycles and reducing experimental workloads. However, current methods struggle with the vast combinatorial space of potential protein edits and cannot explicitly conduct protein editing using biotext instructions, limiting their interactivity with human feedback. To fill these gaps, we propose a novel method called ProtET for efficient CLIP-informed protein editing through multi-modality learning. Our approach comprises two stages: in the pretraining stage, contrastive learning aligns protein-biotext representations encoded by two large language models (LLMs), respectively. Subsequently, during the protein editing stage, the fused features from editing instruction texts and original protein sequences serve as the final editing condition for generating target protein sequences. Comprehensive experiments demonstrated the superiority of ProtET in editing proteins to enhance human-expected functionality across multiple attribute domains, including enzyme catalytic activity, protein stability and antibody specific binding ability. And ProtET improves the state-of-the-art results by a large margin, leading to significant stability improvements of 16.67% and 16.90%. This capability positions ProtET to advance real-world artificial protein editing, potentially addressing unmet academic, industrial, and clinical needs.
Generative AI for Controllable Protein Sequence Design: A Survey
Feb 16, 2024

Abstract:The design of novel protein sequences with targeted functionalities underpins a central theme in protein engineering, impacting diverse fields such as drug discovery and enzymatic engineering. However, navigating this vast combinatorial search space remains a severe challenge due to time and financial constraints. This scenario is rapidly evolving as the transformative advancements in AI, particularly in the realm of generative models and optimization algorithms, have been propelling the protein design field towards an unprecedented revolution. In this survey, we systematically review recent advances in generative AI for controllable protein sequence design. To set the stage, we first outline the foundational tasks in protein sequence design in terms of the constraints involved and present key generative models and optimization algorithms. We then offer in-depth reviews of each design task and discuss the pertinent applications. Finally, we identify the unresolved challenges and highlight research opportunities that merit deeper exploration.
From molecules to scaffolds to functional groups: building context-dependent molecular representation via multi-channel learning
Nov 05, 2023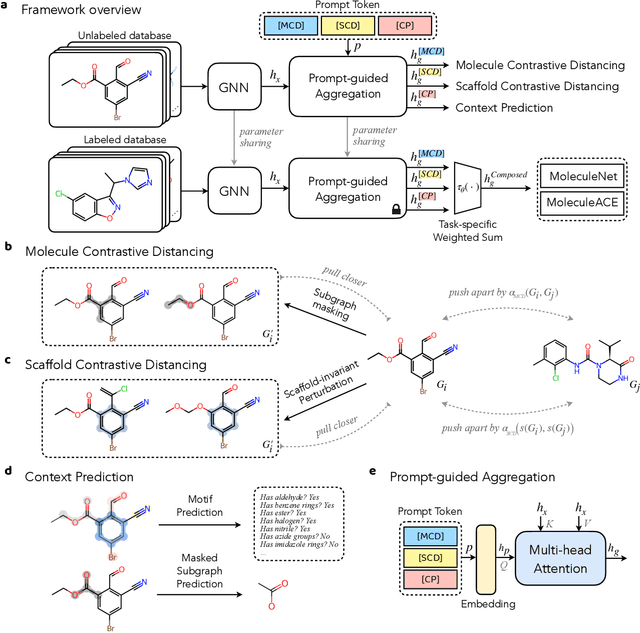
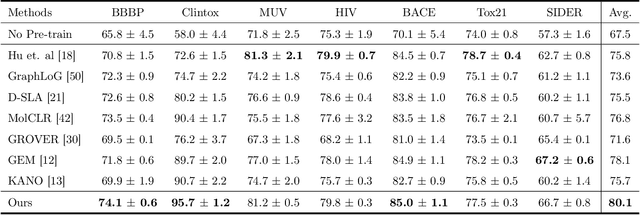
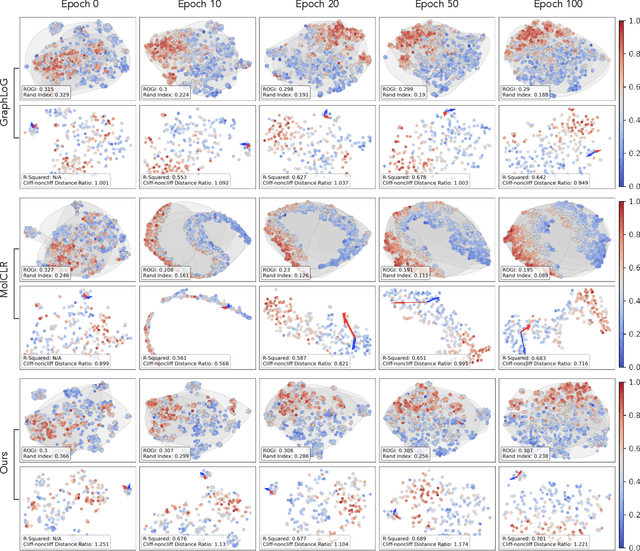

Abstract:Reliable molecular property prediction is essential for various scientific endeavors and industrial applications, such as drug discovery. However, the scarcity of data, combined with the highly non-linear causal relationships between physicochemical and biological properties and conventional molecular featurization schemes, complicates the development of robust molecular machine learning models. Self-supervised learning (SSL) has emerged as a popular solution, utilizing large-scale, unannotated molecular data to learn a foundational representation of chemical space that might be advantageous for downstream tasks. Yet, existing molecular SSL methods largely overlook domain-specific knowledge, such as molecular similarity and scaffold importance, as well as the context of the target application when operating over the large chemical space. This paper introduces a novel learning framework that leverages the knowledge of structural hierarchies within molecular structures, embeds them through separate pre-training tasks over distinct channels, and employs a task-specific channel selection to compose a context-dependent representation. Our approach demonstrates competitive performance across various molecular property benchmarks and establishes some state-of-the-art results. It further offers unprecedented advantages in particularly challenging yet ubiquitous scenarios like activity cliffs with enhanced robustness and generalizability compared to other baselines.
MolHF: A Hierarchical Normalizing Flow for Molecular Graph Generation
May 15, 2023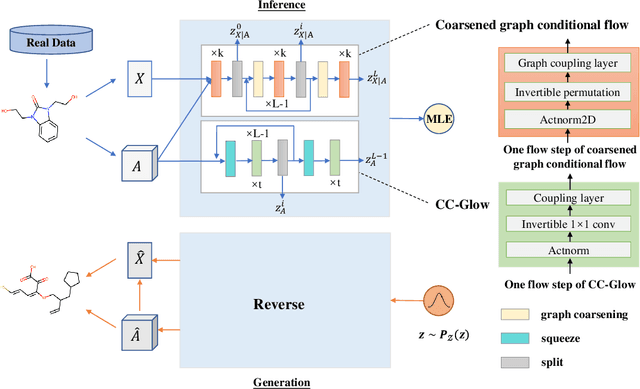
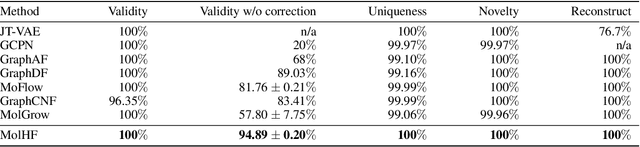
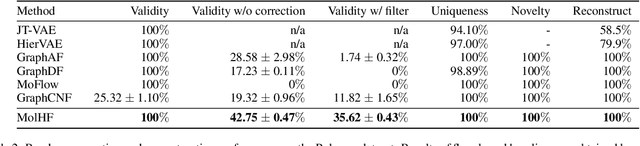
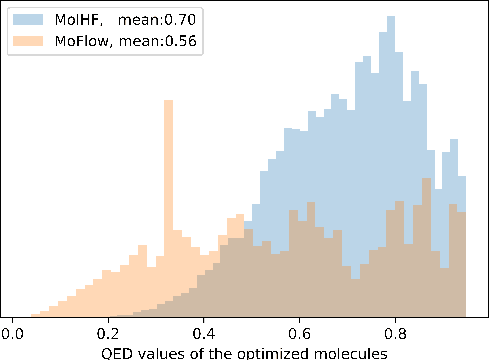
Abstract:Molecular de novo design is a critical yet challenging task in scientific fields, aiming to design novel molecular structures with desired property profiles. Significant progress has been made by resorting to generative models for graphs. However, limited attention is paid to hierarchical generative models, which can exploit the inherent hierarchical structure (with rich semantic information) of the molecular graphs and generate complex molecules of larger size that we shall demonstrate to be difficult for most existing models. The primary challenge to hierarchical generation is the non-differentiable issue caused by the generation of intermediate discrete coarsened graph structures. To sidestep this issue, we cast the tricky hierarchical generation problem over discrete spaces as the reverse process of hierarchical representation learning and propose MolHF, a new hierarchical flow-based model that generates molecular graphs in a coarse-to-fine manner. Specifically, MolHF first generates bonds through a multi-scale architecture, then generates atoms based on the coarsened graph structure at each scale. We demonstrate that MolHF achieves state-of-the-art performance in random generation and property optimization, implying its high capacity to model data distribution. Furthermore, MolHF is the first flow-based model that can be applied to model larger molecules (polymer) with more than 100 heavy atoms. The code and models are available at https://github.com/violet-sto/MolHF.
Sample-efficient Multi-objective Molecular Optimization with GFlowNets
Feb 08, 2023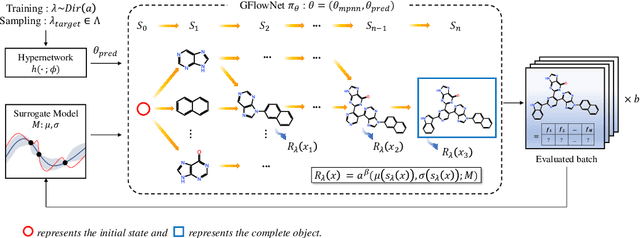
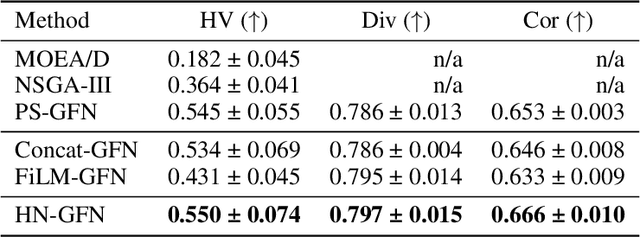
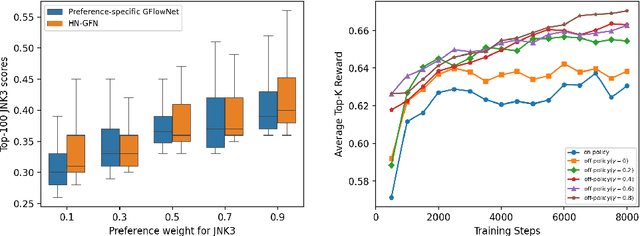
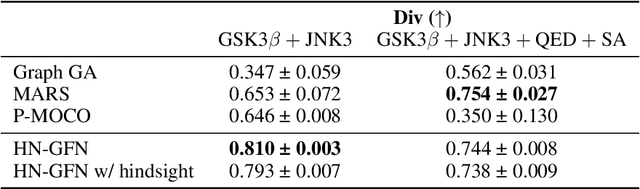
Abstract:Many crucial scientific problems involve designing novel molecules with desired properties, which can be formulated as an expensive black-box optimization problem over the discrete chemical space. Computational methods have achieved initial success but still struggle with simultaneously optimizing multiple competing properties in a sample-efficient manner. In this work, we propose a multi-objective Bayesian optimization (MOBO) algorithm leveraging the hypernetwork-based GFlowNets (HN-GFN) as an acquisition function optimizer, with the purpose of sampling a diverse batch of candidate molecular graphs from an approximate Pareto front. Using a single preference-conditioned hypernetwork, HN-GFN learns to explore various trade-offs between objectives. Inspired by reinforcement learning, we further propose a hindsight-like off-policy strategy to share high-performing molecules among different preferences in order to speed up learning for HN-GFN. Through synthetic experiments, we illustrate that HN-GFN has adequate capacity to generalize over preferences. Extensive experiments show that our framework outperforms the best baselines by a large margin in terms of hypervolume in various real-world MOBO settings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge