Sample-efficient Multi-objective Molecular Optimization with GFlowNets
Paper and Code
Feb 08, 2023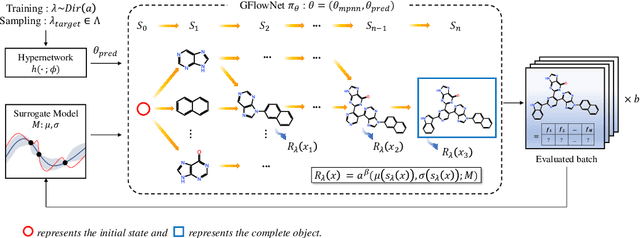
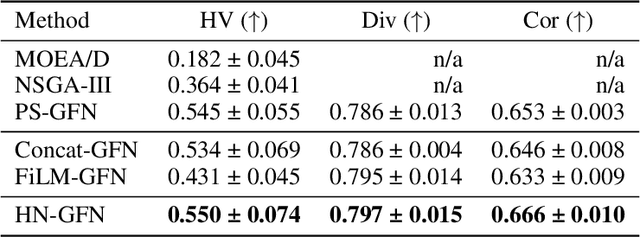
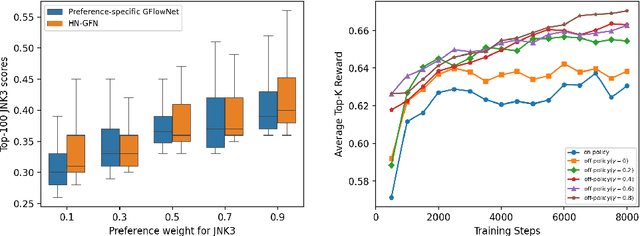
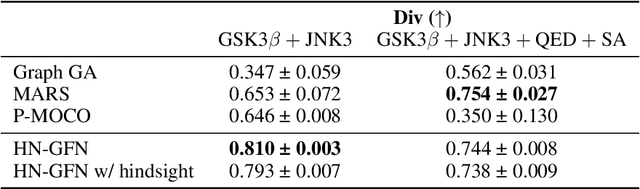
Many crucial scientific problems involve designing novel molecules with desired properties, which can be formulated as an expensive black-box optimization problem over the discrete chemical space. Computational methods have achieved initial success but still struggle with simultaneously optimizing multiple competing properties in a sample-efficient manner. In this work, we propose a multi-objective Bayesian optimization (MOBO) algorithm leveraging the hypernetwork-based GFlowNets (HN-GFN) as an acquisition function optimizer, with the purpose of sampling a diverse batch of candidate molecular graphs from an approximate Pareto front. Using a single preference-conditioned hypernetwork, HN-GFN learns to explore various trade-offs between objectives. Inspired by reinforcement learning, we further propose a hindsight-like off-policy strategy to share high-performing molecules among different preferences in order to speed up learning for HN-GFN. Through synthetic experiments, we illustrate that HN-GFN has adequate capacity to generalize over preferences. Extensive experiments show that our framework outperforms the best baselines by a large margin in terms of hypervolume in various real-world MOBO settings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge