Yuqiang Han
Generative AI for Controllable Protein Sequence Design: A Survey
Feb 16, 2024

Abstract:The design of novel protein sequences with targeted functionalities underpins a central theme in protein engineering, impacting diverse fields such as drug discovery and enzymatic engineering. However, navigating this vast combinatorial search space remains a severe challenge due to time and financial constraints. This scenario is rapidly evolving as the transformative advancements in AI, particularly in the realm of generative models and optimization algorithms, have been propelling the protein design field towards an unprecedented revolution. In this survey, we systematically review recent advances in generative AI for controllable protein sequence design. To set the stage, we first outline the foundational tasks in protein sequence design in terms of the constraints involved and present key generative models and optimization algorithms. We then offer in-depth reviews of each design task and discuss the pertinent applications. Finally, we identify the unresolved challenges and highlight research opportunities that merit deeper exploration.
Contrastive Knowledge Amalgamation for Unsupervised Image Classification
Jul 27, 2023Abstract:Knowledge amalgamation (KA) aims to learn a compact student model to handle the joint objective from multiple teacher models that are are specialized for their own tasks respectively. Current methods focus on coarsely aligning teachers and students in the common representation space, making it difficult for the student to learn the proper decision boundaries from a set of heterogeneous teachers. Besides, the KL divergence in previous works only minimizes the probability distribution difference between teachers and the student, ignoring the intrinsic characteristics of teachers. Therefore, we propose a novel Contrastive Knowledge Amalgamation (CKA) framework, which introduces contrastive losses and an alignment loss to achieve intra-class cohesion and inter-class separation.Contrastive losses intra- and inter- models are designed to widen the distance between representations of different classes. The alignment loss is introduced to minimize the sample-level distribution differences of teacher-student models in the common representation space.Furthermore, the student learns heterogeneous unsupervised classification tasks through soft targets efficiently and flexibly in the task-level amalgamation. Extensive experiments on benchmarks demonstrate the generalization capability of CKA in the amalgamation of specific task as well as multiple tasks. Comprehensive ablation studies provide a further insight into our CKA.
Deep Reinforcement Learning for Entity Alignment
Mar 07, 2022
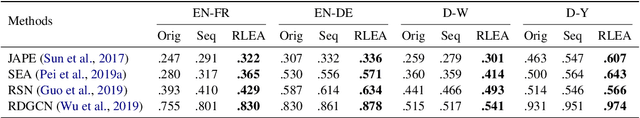


Abstract:Embedding-based methods have attracted increasing attention in recent entity alignment (EA) studies. Although great promise they can offer, there are still several limitations. The most notable is that they identify the aligned entities based on cosine similarity, ignoring the semantics underlying the embeddings themselves. Furthermore, these methods are shortsighted, heuristically selecting the closest entity as the target and allowing multiple entities to match the same candidate. To address these limitations, we model entity alignment as a sequential decision-making task, in which an agent sequentially decides whether two entities are matched or mismatched based on their representation vectors. The proposed reinforcement learning (RL)-based entity alignment framework can be flexibly adapted to most embedding-based EA methods. The experimental results demonstrate that it consistently advances the performance of several state-of-the-art methods, with a maximum improvement of 31.1% on Hits@1.
DialMed: A Dataset for Dialogue-based Medication Recommendation
Feb 22, 2022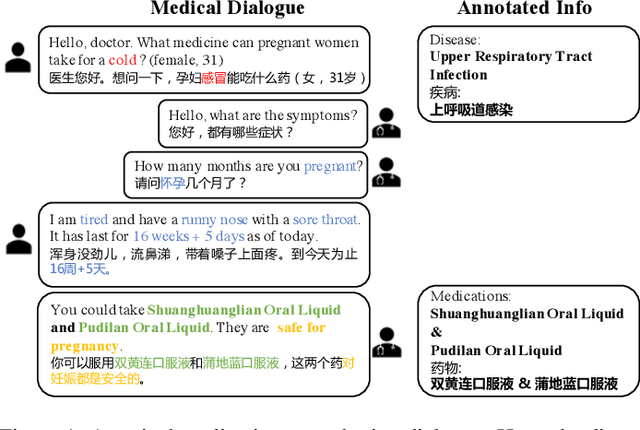

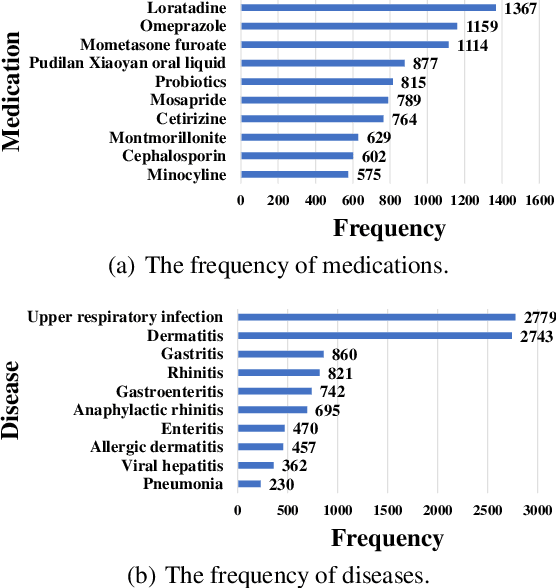

Abstract:Medication recommendation is a crucial task for intelligent healthcare systems. Previous studies mainly recommend medications with electronic health records(EHRs). However, some details of interactions between doctors and patients may be ignored in EHRs, which are essential for automatic medication recommendation. Therefore, we make the first attempt to recommend medications with the conversations between doctors and patients. In this work, we construct DialMed, the first high-quality dataset for medical dialogue-based medication recommendation task. It contains 11,996 medical dialogues related to 16 common diseases from 3 departments and 70 corresponding common medications. Furthermore, we propose a Dialogue structure and Disease knowledge aware Network(DDN), where a graph attention network is utilized to model the dialogue structure and the knowledge graph is used to introduce external disease knowledge. The extensive experimental results demonstrate that the proposed method is a promising solution to recommend medications with medical dialogues. The dataset and code are available at https://github.com/Hhhhhhhzf/DialMed.
Prompt-Guided Injection of Conformation to Pre-trained Protein Model
Feb 07, 2022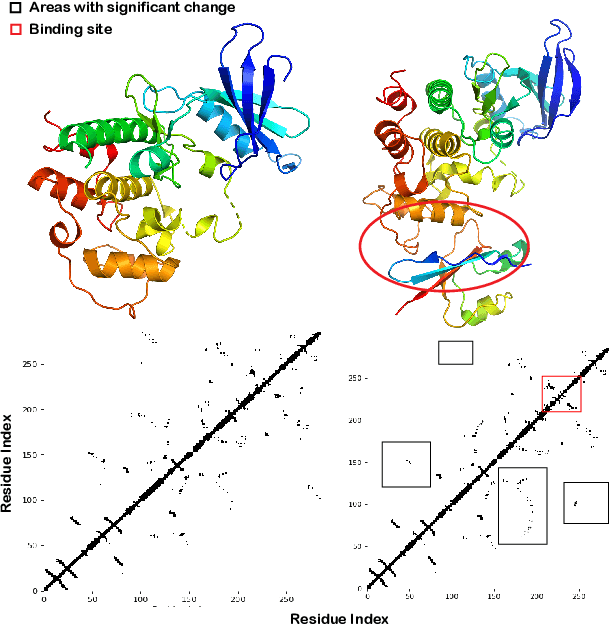
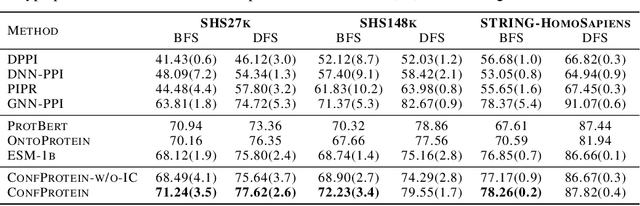
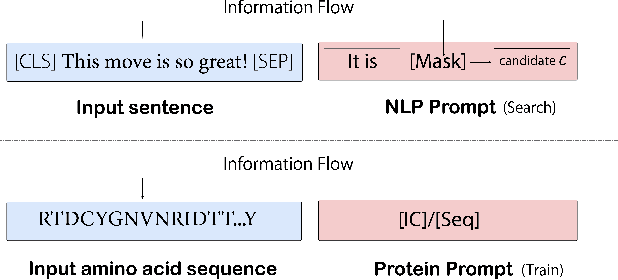

Abstract:Pre-trained protein models (PTPMs) represent a protein with one fixed embedding and thus are not capable for diverse tasks. For example, protein structures can shift, namely protein folding, between several conformations in various biological processes. To enable PTPMs to produce task-aware representations, we propose to learn interpretable, pluggable and extensible protein prompts as a way of injecting task-related knowledge into PTPMs. In this regard, prior PTPM optimization with the masked language modeling task can be interpreted as learning a sequence prompt (Seq prompt) that enables PTPMs to capture the sequential dependency between amino acids. To incorporate conformational knowledge to PTPMs, we propose an interaction-conformation prompt (IC prompt) that is learned through back-propagation with the protein-protein interaction task. As an instantiation, we present a conformation-aware pre-trained protein model that learns both sequence and interaction-conformation prompts in a multi-task setting. We conduct comprehensive experiments on nine protein datasets. Results confirm our expectation that using the sequence prompt does not hurt PTPMs' performance on sequence-related tasks while incorporating the interaction-conformation prompt significantly improves PTPMs' performance on tasks where conformational knowledge counts. We also show the learned prompts can be combined and extended to deal with new complex tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge