Huajun Chen
Zhejiang University
From Data to Behavior: Predicting Unintended Model Behaviors Before Training
Feb 04, 2026Abstract:Large Language Models (LLMs) can acquire unintended biases from seemingly benign training data even without explicit cues or malicious content. Existing methods struggle to detect such risks before fine-tuning, making post hoc evaluation costly and inefficient. To address this challenge, we introduce Data2Behavior, a new task for predicting unintended model behaviors prior to training. We also propose Manipulating Data Features (MDF), a lightweight approach that summarizes candidate data through their mean representations and injects them into the forward pass of a base model, allowing latent statistical signals in the data to shape model activations and reveal potential biases and safety risks without updating any parameters. MDF achieves reliable prediction while consuming only about 20% of the GPU resources required for fine-tuning. Experiments on Qwen3-14B, Qwen2.5-32B-Instruct, and Gemma-3-12b-it confirm that MDF can anticipate unintended behaviors and provide insight into pre-training vulnerabilities.
Why Steering Works: Toward a Unified View of Language Model Parameter Dynamics
Feb 02, 2026Abstract:Methods for controlling large language models (LLMs), including local weight fine-tuning, LoRA-based adaptation, and activation-based interventions, are often studied in isolation, obscuring their connections and making comparison difficult. In this work, we present a unified view that frames these interventions as dynamic weight updates induced by a control signal, placing them within a single conceptual framework. Building on this view, we propose a unified preference-utility analysis that separates control effects into preference, defined as the tendency toward a target concept, and utility, defined as coherent and task-valid generation, and measures both on a shared log-odds scale using polarity-paired contrastive examples. Across methods, we observe a consistent trade-off between preference and utility: stronger control increases preference while predictably reducing utility. We further explain this behavior through an activation manifold perspective, in which control shifts representations along target-concept directions to enhance preference, while utility declines primarily when interventions push representations off the model's valid-generation manifold. Finally, we introduce a new steering approach SPLIT guided by this analysis that improves preference while better preserving utility. Code is available at https://github.com/zjunlp/EasyEdit/blob/main/examples/SPLIT.md.
Temp-R1: A Unified Autonomous Agent for Complex Temporal KGQA via Reverse Curriculum Reinforcement Learning
Jan 26, 2026Abstract:Temporal Knowledge Graph Question Answering (TKGQA) is inherently challenging, as it requires sophisticated reasoning over dynamic facts with multi-hop dependencies and complex temporal constraints. Existing methods rely on fixed workflows and expensive closed-source APIs, limiting flexibility and scalability. We propose Temp-R1, the first autonomous end-to-end agent for TKGQA trained through reinforcement learning. To address cognitive overload in single-action reasoning, we expand the action space with specialized internal actions alongside external action. To prevent shortcut learning on simple questions, we introduce reverse curriculum learning that trains on difficult questions first, forcing the development of sophisticated reasoning before transferring to easier cases. Our 8B-parameter Temp-R1 achieves state-of-the-art performance on MultiTQ and TimelineKGQA, improving 19.8% over strong baselines on complex questions. Our work establishes a new paradigm for autonomous temporal reasoning agents. Our code will be publicly available soon at https://github.com/zjukg/Temp-R1.
Aligning Agentic World Models via Knowledgeable Experience Learning
Jan 19, 2026Abstract:Current Large Language Models (LLMs) exhibit a critical modal disconnect: they possess vast semantic knowledge but lack the procedural grounding to respect the immutable laws of the physical world. Consequently, while these agents implicitly function as world models, their simulations often suffer from physical hallucinations-generating plans that are logically sound but physically unexecutable. Existing alignment strategies predominantly rely on resource-intensive training or fine-tuning, which attempt to compress dynamic environmental rules into static model parameters. However, such parametric encapsulation is inherently rigid, struggling to adapt to the open-ended variability of physical dynamics without continuous, costly retraining. To bridge this gap, we introduce WorldMind, a framework that autonomously constructs a symbolic World Knowledge Repository by synthesizing environmental feedback. Specifically, it unifies Process Experience to enforce physical feasibility via prediction errors and Goal Experience to guide task optimality through successful trajectories. Experiments on EB-ALFRED and EB-Habitat demonstrate that WorldMind achieves superior performance compared to baselines with remarkable cross-model and cross-environment transferability.
CoG: Controllable Graph Reasoning via Relational Blueprints and Failure-Aware Refinement over Knowledge Graphs
Jan 16, 2026Abstract:Large Language Models (LLMs) have demonstrated remarkable reasoning capabilities but often grapple with reliability challenges like hallucinations. While Knowledge Graphs (KGs) offer explicit grounding, existing paradigms of KG-augmented LLMs typically exhibit cognitive rigidity--applying homogeneous search strategies that render them vulnerable to instability under neighborhood noise and structural misalignment leading to reasoning stagnation. To address these challenges, we propose CoG, a training-free framework inspired by Dual-Process Theory that mimics the interplay between intuition and deliberation. First, functioning as the fast, intuitive process, the Relational Blueprint Guidance module leverages relational blueprints as interpretable soft structural constraints to rapidly stabilize the search direction against noise. Second, functioning as the prudent, analytical process, the Failure-Aware Refinement module intervenes upon encountering reasoning impasses. It triggers evidence-conditioned reflection and executes controlled backtracking to overcome reasoning stagnation. Experimental results on three benchmarks demonstrate that CoG significantly outperforms state-of-the-art approaches in both accuracy and efficiency.
Can We Predict Before Executing Machine Learning Agents?
Jan 09, 2026Abstract:Autonomous machine learning agents have revolutionized scientific discovery, yet they remain constrained by a Generate-Execute-Feedback paradigm. Previous approaches suffer from a severe Execution Bottleneck, as hypothesis evaluation relies strictly on expensive physical execution. To bypass these physical constraints, we internalize execution priors to substitute costly runtime checks with instantaneous predictive reasoning, drawing inspiration from World Models. In this work, we formalize the task of Data-centric Solution Preference and construct a comprehensive corpus of 18,438 pairwise comparisons. We demonstrate that LLMs exhibit significant predictive capabilities when primed with a Verified Data Analysis Report, achieving 61.5% accuracy and robust confidence calibration. Finally, we instantiate this framework in FOREAGENT, an agent that employs a Predict-then-Verify loop, achieving a 6x acceleration in convergence while surpassing execution-based baselines by +6%. Our code and dataset will be publicly available soon at https://github.com/zjunlp/predict-before-execute.
Illusions of Confidence? Diagnosing LLM Truthfulness via Neighborhood Consistency
Jan 09, 2026Abstract:As Large Language Models (LLMs) are increasingly deployed in real-world settings, correctness alone is insufficient. Reliable deployment requires maintaining truthful beliefs under contextual perturbations. Existing evaluations largely rely on point-wise confidence like Self-Consistency, which can mask brittle belief. We show that even facts answered with perfect self-consistency can rapidly collapse under mild contextual interference. To address this gap, we propose Neighbor-Consistency Belief (NCB), a structural measure of belief robustness that evaluates response coherence across a conceptual neighborhood. To validate the efficiency of NCB, we introduce a new cognitive stress-testing protocol that probes outputs stability under contextual interference. Experiments across multiple LLMs show that the performance of high-NCB data is relatively more resistant to interference. Finally, we present Structure-Aware Training (SAT), which optimizes context-invariant belief structure and reduces long-tail knowledge brittleness by approximately 30%. Code will be available at https://github.com/zjunlp/belief.
ClinDEF: A Dynamic Evaluation Framework for Large Language Models in Clinical Reasoning
Dec 29, 2025Abstract:Clinical diagnosis begins with doctor-patient interaction, during which physicians iteratively gather information, determine examination and refine differential diagnosis through patients' response. This dynamic clinical-reasoning process is poorly represented by existing LLM benchmarks that focus on static question-answering. To mitigate these gaps, recent methods explore dynamic medical frameworks involving interactive clinical dialogues. Although effective, they often rely on limited, contamination-prone datasets and lack granular, multi-level evaluation. In this work, we propose ClinDEF, a dynamic framework for assessing clinical reasoning in LLMs through simulated diagnostic dialogues. Grounded in a disease knowledge graph, our method dynamically generates patient cases and facilitates multi-turn interactions between an LLM-based doctor and an automated patient agent. Our evaluation protocol goes beyond diagnostic accuracy by incorporating fine-grained efficiency analysis and rubric-based assessment of diagnostic quality. Experiments show that ClinDEF effectively exposes critical clinical reasoning gaps in state-of-the-art LLMs, offering a more nuanced and clinically meaningful evaluation paradigm.
Retrieval-augmented Prompt Learning for Pre-trained Foundation Models
Dec 23, 2025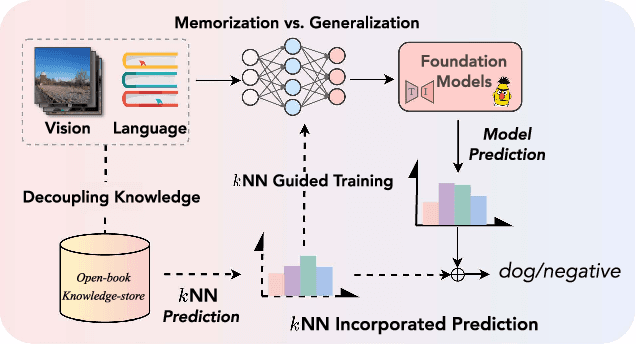
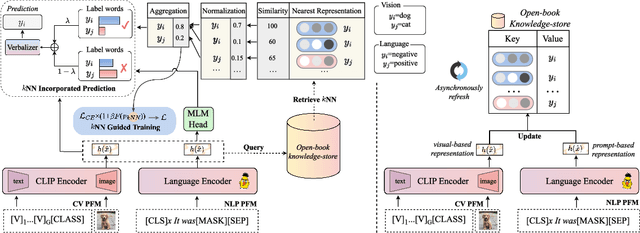
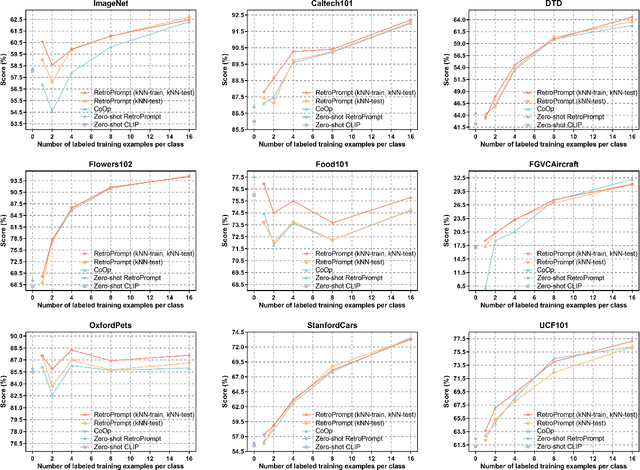
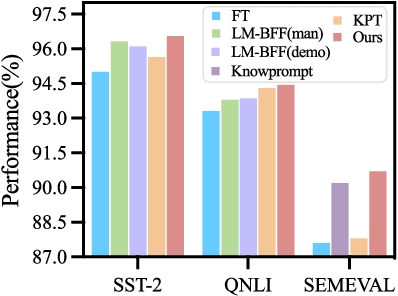
Abstract:The pre-trained foundation models (PFMs) have become essential for facilitating large-scale multimodal learning. Researchers have effectively employed the ``pre-train, prompt, and predict'' paradigm through prompt learning to induce improved few-shot performance. However, prompt learning approaches for PFMs still follow a parametric learning paradigm. As such, the stability of generalization in memorization and rote learning can be compromised. More specifically, conventional prompt learning might face difficulties in fully utilizing atypical instances and avoiding overfitting to shallow patterns with limited data during the process of fully-supervised training. To overcome these constraints, we present our approach, named RetroPrompt, which aims to achieve a balance between memorization and generalization by decoupling knowledge from mere memorization. Unlike traditional prompting methods, RetroPrompt leverages a publicly accessible knowledge base generated from the training data and incorporates a retrieval mechanism throughout the input, training, and inference stages. This enables the model to actively retrieve relevant contextual information from the corpus, thereby enhancing the available cues. We conduct comprehensive experiments on a variety of datasets across natural language processing and computer vision tasks to demonstrate the superior performance of our proposed approach, RetroPrompt, in both zero-shot and few-shot scenarios. Through detailed analysis of memorization patterns, we observe that RetroPrompt effectively reduces the reliance on rote memorization, leading to enhanced generalization.
Thinker: Training LLMs in Hierarchical Thinking for Deep Search via Multi-Turn Interaction
Nov 14, 2025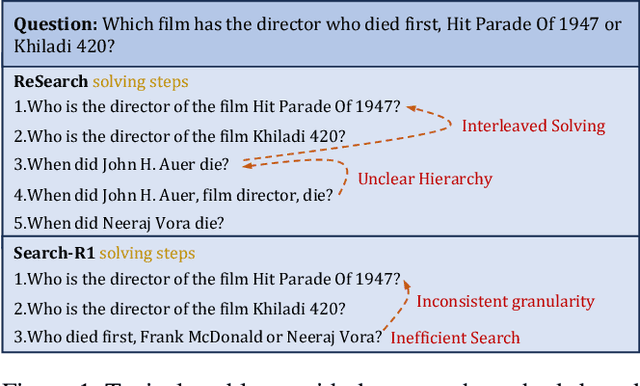
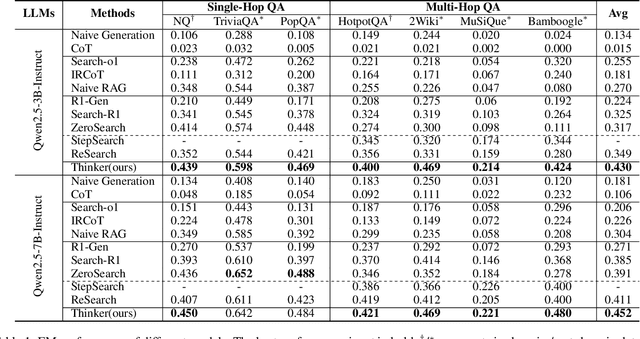
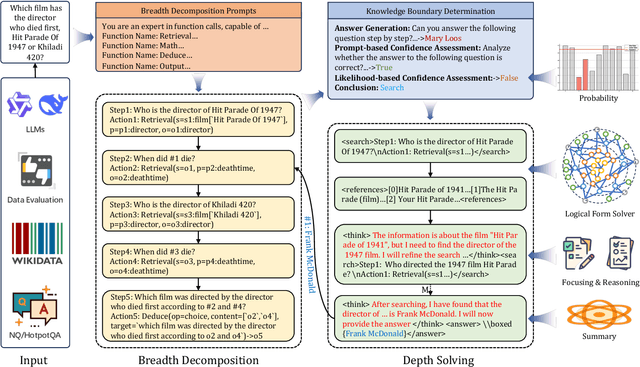
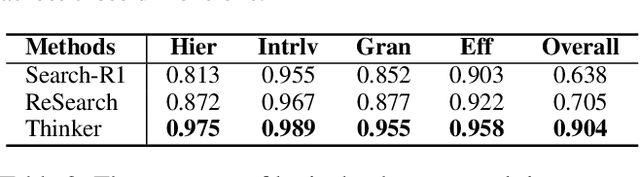
Abstract:Efficient retrieval of external knowledge bases and web pages is crucial for enhancing the reasoning abilities of LLMs. Previous works on training LLMs to leverage external retrievers for solving complex problems have predominantly employed end-to-end reinforcement learning. However, these approaches neglect supervision over the reasoning process, making it difficult to guarantee logical coherence and rigor. To address these limitations, we propose Thinker, a hierarchical thinking model for deep search through multi-turn interaction, making the reasoning process supervisable and verifiable. It decomposes complex problems into independently solvable sub-problems, each dually represented in both natural language and an equivalent logical function to support knowledge base and web searches. Concurrently, dependencies between sub-problems are passed as parameters via these logical functions, enhancing the logical coherence of the problem-solving process. To avoid unnecessary external searches, we perform knowledge boundary determination to check if a sub-problem is within the LLM's intrinsic knowledge, allowing it to answer directly. Experimental results indicate that with as few as several hundred training samples, the performance of Thinker is competitive with established baselines. Furthermore, when scaled to the full training set, Thinker significantly outperforms these methods across various datasets and model sizes. The source code is available at https://github.com/OpenSPG/KAG-Thinker.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge