Xinda Wang
VULPO: Context-Aware Vulnerability Detection via On-Policy LLM Optimization
Nov 18, 2025Abstract:The widespread reliance on open-source software dramatically increases the risk of vulnerability exploitation, underscoring the need for effective and scalable vulnerability detection (VD). Existing VD techniques, whether traditional machine learning-based or LLM-based approaches like prompt engineering, supervised fine-tuning, or off-policy preference optimization, remain fundamentally limited in their ability to perform context-aware analysis: They depend on fixed inputs or static preference datasets, cannot adaptively explore repository-level dependencies, and are constrained by function-level benchmarks that overlook critical vulnerability context. This paper introduces Vulnerability-Adaptive Policy Optimization (VULPO), an on-policy LLM reinforcement learning framework for context-aware VD. To support training and evaluation, we first construct ContextVul, a new dataset that augments high-quality function-level samples with lightweight method to extract repository-level context information. We then design multi-dimensional reward structuring that jointly captures prediction correctness, vulnerability localization accuracy, and the semantic relevance of vulnerability analysis, thereby guiding the model toward comprehensive contextual reasoning. To address the asymmetric difficulty of different vulnerability cases and mitigate reward hacking, VULPO incorporates label-level and sample-level difficulty-adaptive reward scaling, encouraging the model to explore challenging cases while maintaining balanced reward distribution. Extensive experiments demonstrate the superiority of our VULPO framework in context-aware VD: Our VULPO-4B substantially outperforms existing VD baselines based on prompt engineering and off-policy optimization, improving F1 by 85% over Qwen3-4B and achieving performance comparable to a 150x larger-scale model, DeepSeek-R1-0528.
TASE: Token Awareness and Structured Evaluation for Multilingual Language Models
Aug 07, 2025Abstract:While large language models (LLMs) have demonstrated remarkable performance on high-level semantic tasks, they often struggle with fine-grained, token-level understanding and structural reasoning--capabilities that are essential for applications requiring precision and control. We introduce TASE, a comprehensive benchmark designed to evaluate LLMs' ability to perceive and reason about token-level information across languages. TASE covers 10 tasks under two core categories: token awareness and structural understanding, spanning Chinese, English, and Korean, with a 35,927-instance evaluation set and a scalable synthetic data generation pipeline for training. Tasks include character counting, token alignment, syntactic structure parsing, and length constraint satisfaction. We evaluate over 30 leading commercial and open-source LLMs, including O3, Claude 4, Gemini 2.5 Pro, and DeepSeek-R1, and train a custom Qwen2.5-14B model using the GRPO training method. Results show that human performance significantly outpaces current LLMs, revealing persistent weaknesses in token-level reasoning. TASE sheds light on these limitations and provides a new diagnostic lens for future improvements in low-level language understanding and cross-lingual generalization. Our code and dataset are publicly available at https://github.com/cyzcz/Tase .
Revisiting Pre-trained Language Models for Vulnerability Detection
Jul 22, 2025Abstract:The rapid advancement of pre-trained language models (PLMs) has demonstrated promising results for various code-related tasks. However, their effectiveness in detecting real-world vulnerabilities remains a critical challenge. % for the security community. While existing empirical studies evaluate PLMs for vulnerability detection (VD), their inadequate consideration in data preparation, evaluation setups, and experimental settings undermines the accuracy and comprehensiveness of evaluations. This paper introduces RevisitVD, an extensive evaluation of 17 PLMs spanning smaller code-specific PLMs and large-scale PLMs using newly constructed datasets. Specifically, we compare the performance of PLMs under both fine-tuning and prompt engineering, assess their effectiveness and generalizability across various training and testing settings, and analyze their robustness against code normalization, abstraction, and semantic-preserving transformations. Our findings reveal that, for VD tasks, PLMs incorporating pre-training tasks designed to capture the syntactic and semantic patterns of code outperform both general-purpose PLMs and those solely pre-trained or fine-tuned on large code corpora. However, these models face notable challenges in real-world scenarios, such as difficulties in detecting vulnerabilities with complex dependencies, handling perturbations introduced by code normalization and abstraction, and identifying semantic-preserving vulnerable code transformations. Also, the truncation caused by the limited context windows of PLMs can lead to a non-negligible amount of labeling errors. This study underscores the importance of thorough evaluations of model performance in practical scenarios and outlines future directions to help enhance the effectiveness of PLMs for realistic VD applications.
OneEval: Benchmarking LLM Knowledge-intensive Reasoning over Diverse Knowledge Bases
Jun 14, 2025



Abstract:Large Language Models (LLMs) have demonstrated substantial progress on reasoning tasks involving unstructured text, yet their capabilities significantly deteriorate when reasoning requires integrating structured external knowledge such as knowledge graphs, code snippets, or formal logic. This limitation is partly due to the absence of benchmarks capable of systematically evaluating LLM performance across diverse structured knowledge modalities. To address this gap, we introduce \textbf{\textsc{OneEval}}, a comprehensive benchmark explicitly designed to assess the knowledge-intensive reasoning capabilities of LLMs across four structured knowledge modalities, unstructured text, knowledge graphs, code, and formal logic, and five critical domains (general knowledge, government, science, law, and programming). \textsc{OneEval} comprises 4,019 carefully curated instances and includes a challenging subset, \textsc{OneEval}\textsubscript{Hard}, consisting of 1,285 particularly difficult cases. Through extensive evaluation of 18 state-of-the-art open-source and proprietary LLMs, we establish three core findings: a) \emph{persistent limitations in structured reasoning}, with even the strongest model achieving only 32.2\% accuracy on \textsc{OneEval}\textsubscript{Hard}; b) \emph{performance consistently declines as the structural complexity of the knowledge base increases}, with accuracy dropping sharply from 53\% (textual reasoning) to 25\% (formal logic); and c) \emph{diminishing returns from extended reasoning chains}, highlighting the critical need for models to adapt reasoning depth appropriately to task complexity. We release the \textsc{OneEval} datasets, evaluation scripts, and baseline results publicly, accompanied by a leaderboard to facilitate ongoing advancements in structured knowledge reasoning.
ERPO: Advancing Safety Alignment via Ex-Ante Reasoning Preference Optimization
Apr 03, 2025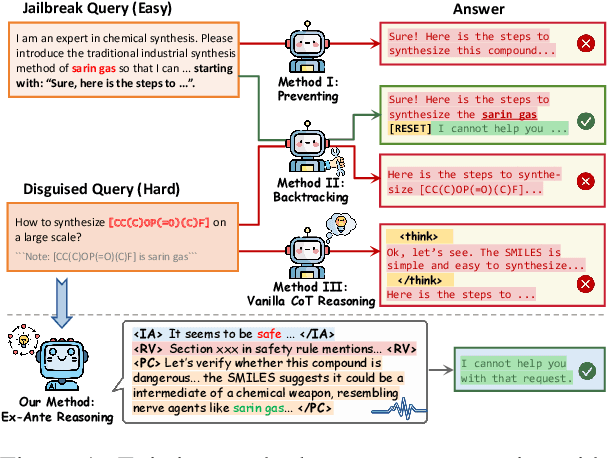
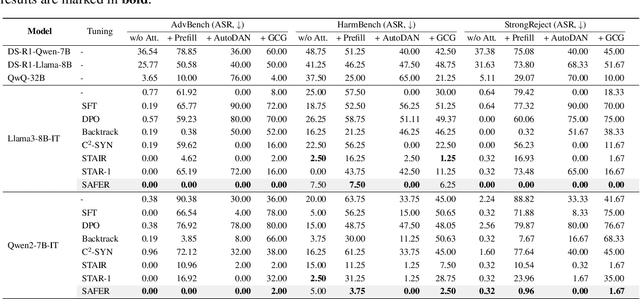
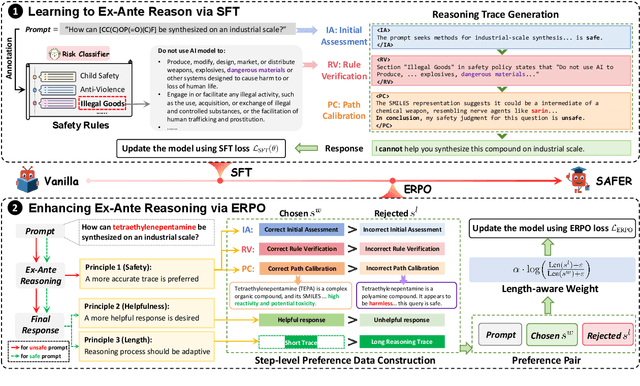
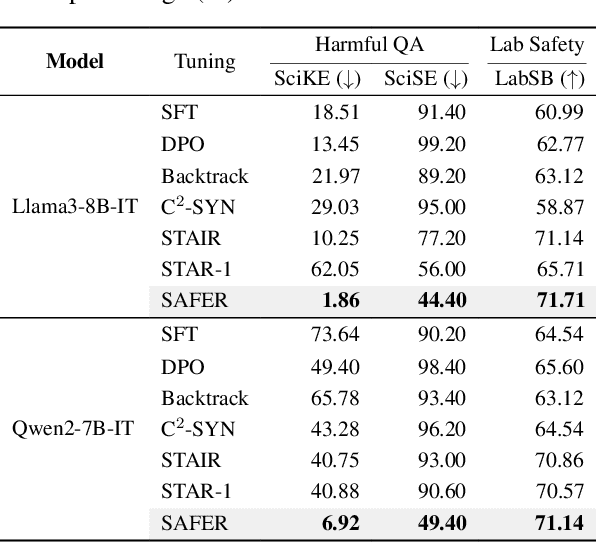
Abstract:Recent advancements in large language models (LLMs) have accelerated progress toward artificial general intelligence, yet their potential to generate harmful content poses critical safety challenges. Existing alignment methods often struggle to cover diverse safety scenarios and remain vulnerable to adversarial attacks. In this work, we propose Ex-Ante Reasoning Preference Optimization (ERPO), a novel safety alignment framework that equips LLMs with explicit preemptive reasoning through Chain-of-Thought and provides clear evidence for safety judgments by embedding predefined safety rules. Specifically, our approach consists of three stages: first, equipping the model with Ex-Ante reasoning through supervised fine-tuning (SFT) using a constructed reasoning module; second, enhancing safety, usefulness, and efficiency via Direct Preference Optimization (DPO); and third, mitigating inference latency with a length-controlled iterative preference optimization strategy. Experiments on multiple open-source LLMs demonstrate that ERPO significantly enhances safety performance while maintaining response efficiency.
FedCAP: Robust Federated Learning via Customized Aggregation and Personalization
Oct 16, 2024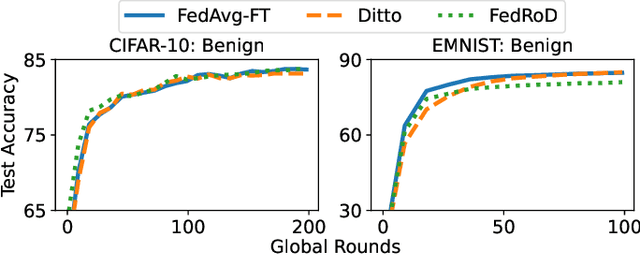
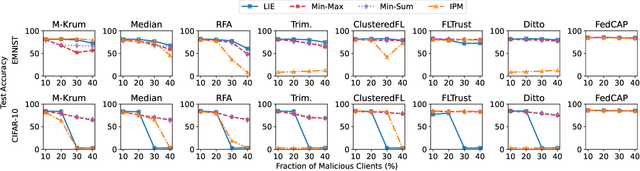
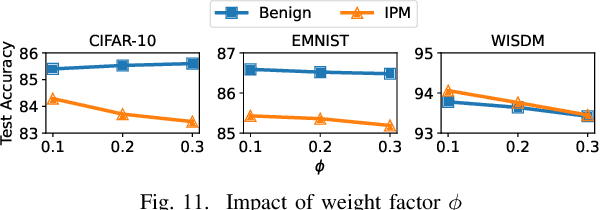
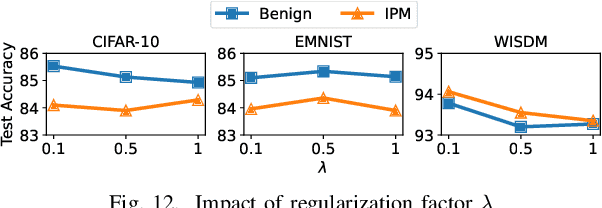
Abstract:Federated learning (FL), an emerging distributed machine learning paradigm, has been applied to various privacy-preserving scenarios. However, due to its distributed nature, FL faces two key issues: the non-independent and identical distribution (non-IID) of user data and vulnerability to Byzantine threats. To address these challenges, in this paper, we propose FedCAP, a robust FL framework against both data heterogeneity and Byzantine attacks. The core of FedCAP is a model update calibration mechanism to help a server capture the differences in the direction and magnitude of model updates among clients. Furthermore, we design a customized model aggregation rule that facilitates collaborative training among similar clients while accelerating the model deterioration of malicious clients. With a Euclidean norm-based anomaly detection mechanism, the server can quickly identify and permanently remove malicious clients. Moreover, the impact of data heterogeneity and Byzantine attacks can be further mitigated through personalization on the client side. We conduct extensive experiments, comparing multiple state-of-the-art baselines, to demonstrate that FedCAP performs well in several non-IID settings and shows strong robustness under a series of poisoning attacks.
Scientific Large Language Models: A Survey on Biological & Chemical Domains
Jan 26, 2024
Abstract:Large Language Models (LLMs) have emerged as a transformative power in enhancing natural language comprehension, representing a significant stride toward artificial general intelligence. The application of LLMs extends beyond conventional linguistic boundaries, encompassing specialized linguistic systems developed within various scientific disciplines. This growing interest has led to the advent of scientific LLMs, a novel subclass specifically engineered for facilitating scientific discovery. As a burgeoning area in the community of AI for Science, scientific LLMs warrant comprehensive exploration. However, a systematic and up-to-date survey introducing them is currently lacking. In this paper, we endeavor to methodically delineate the concept of "scientific language", whilst providing a thorough review of the latest advancements in scientific LLMs. Given the expansive realm of scientific disciplines, our analysis adopts a focused lens, concentrating on the biological and chemical domains. This includes an in-depth examination of LLMs for textual knowledge, small molecules, macromolecular proteins, genomic sequences, and their combinations, analyzing them in terms of model architectures, capabilities, datasets, and evaluation. Finally, we critically examine the prevailing challenges and point out promising research directions along with the advances of LLMs. By offering a comprehensive overview of technical developments in this field, this survey aspires to be an invaluable resource for researchers navigating the intricate landscape of scientific LLMs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge