Haoran Yu
Enhancing Healthcare through Large Language Models: A Study on Medical Question Answering
Aug 08, 2024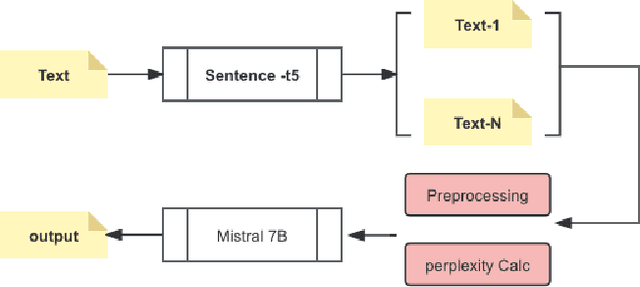
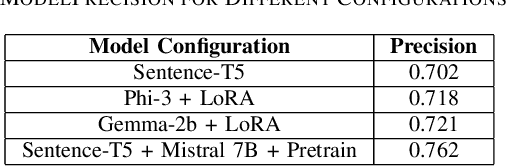
Abstract:In recent years, the application of Large Language Models (LLMs) in healthcare has shown significant promise in improving the accessibility and dissemination of medical knowledge. This paper presents a detailed study of various LLMs trained on the MedQuAD medical question-answering dataset, with a focus on identifying the most effective model for providing accurate medical information. Among the models tested, the Sentence-t5 combined with Mistral 7B demonstrated superior performance, achieving a precision score of 0.762. This model's enhanced capabilities are attributed to its advanced pretraining techniques, robust architecture, and effective prompt construction methodologies. By leveraging these strengths, the Sentence-t5 + Mistral 7B model excels in understanding and generating precise medical answers. Our findings highlight the potential of integrating sophisticated LLMs in medical contexts to facilitate efficient and accurate medical knowledge retrieval, thus significantly enhancing patient education and support.
Prompt-Guided Injection of Conformation to Pre-trained Protein Model
Feb 07, 2022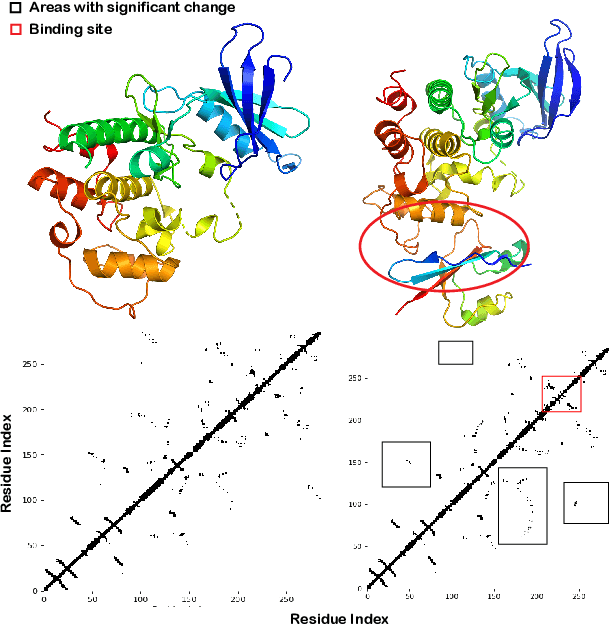
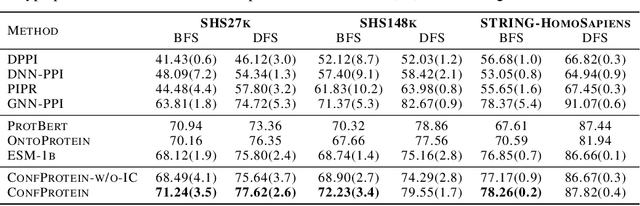
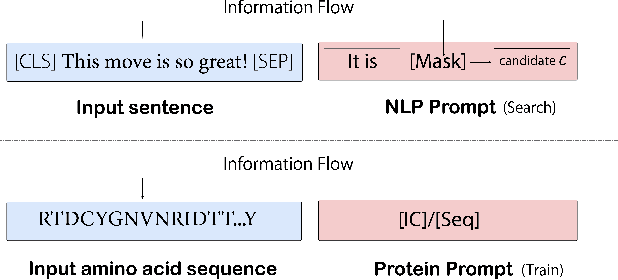

Abstract:Pre-trained protein models (PTPMs) represent a protein with one fixed embedding and thus are not capable for diverse tasks. For example, protein structures can shift, namely protein folding, between several conformations in various biological processes. To enable PTPMs to produce task-aware representations, we propose to learn interpretable, pluggable and extensible protein prompts as a way of injecting task-related knowledge into PTPMs. In this regard, prior PTPM optimization with the masked language modeling task can be interpreted as learning a sequence prompt (Seq prompt) that enables PTPMs to capture the sequential dependency between amino acids. To incorporate conformational knowledge to PTPMs, we propose an interaction-conformation prompt (IC prompt) that is learned through back-propagation with the protein-protein interaction task. As an instantiation, we present a conformation-aware pre-trained protein model that learns both sequence and interaction-conformation prompts in a multi-task setting. We conduct comprehensive experiments on nine protein datasets. Results confirm our expectation that using the sequence prompt does not hurt PTPMs' performance on sequence-related tasks while incorporating the interaction-conformation prompt significantly improves PTPMs' performance on tasks where conformational knowledge counts. We also show the learned prompts can be combined and extended to deal with new complex tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge