Qi Chang
Multi-Scale Cross-Fusion and Edge-Supervision Network for Image Splicing Localization
Dec 17, 2024



Abstract:Image Splicing Localization (ISL) is a fundamental yet challenging task in digital forensics. Although current approaches have achieved promising performance, the edge information is insufficiently exploited, resulting in poor integrality and high false alarms. To tackle this problem, we propose a multi-scale cross-fusion and edge-supervision network for ISL. Specifically, our framework consists of three key steps: multi-scale features cross-fusion, edge mask prediction and edge-supervision localization. Firstly, we input the RGB image and its noise image into a segmentation network to learn multi-scale features, which are then aggregated via a cross-scale fusion followed by a cross-domain fusion to enhance feature representation. Secondly, we design an edge mask prediction module to effectively mine the reliable boundary artifacts. Finally, the cross-fused features and the reliable edge mask information are seamlessly integrated via an attention mechanism to incrementally supervise and facilitate model training. Extensive experiments on publicly available datasets demonstrate that our proposed method is superior to state-of-the-art schemes.
RICAU-Net: Residual-block Inspired Coordinate Attention U-Net for Segmentation of Small and Sparse Calcium Lesions in Cardiac CT
Sep 11, 2024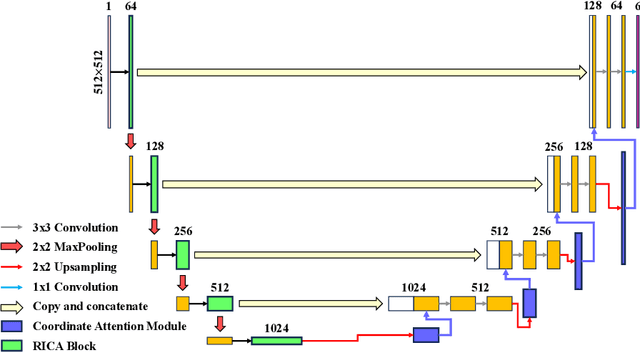
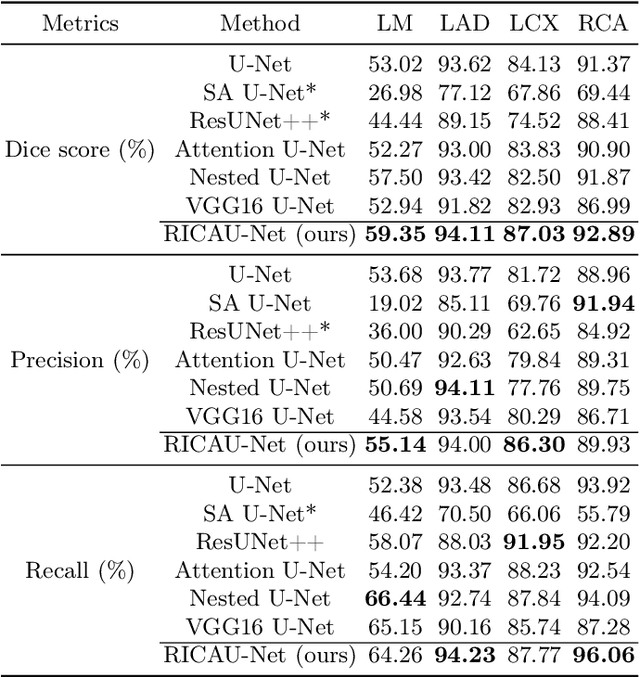
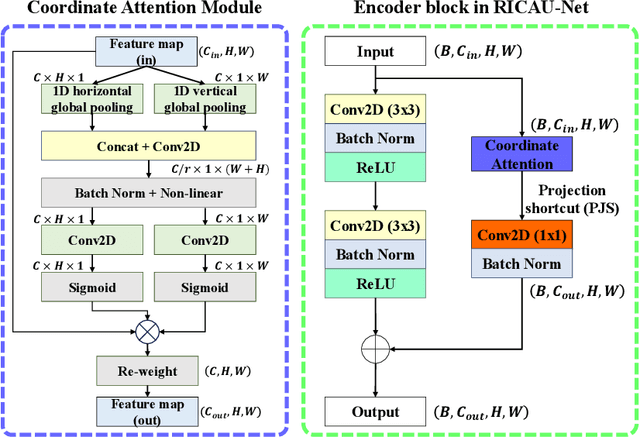
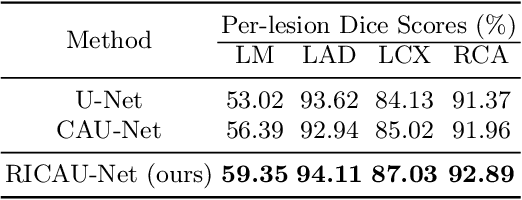
Abstract:The Agatston score, which is the sum of the calcification in the four main coronary arteries, has been widely used in the diagnosis of coronary artery disease (CAD). However, many studies have emphasized the importance of the vessel-specific Agatston score, as calcification in a specific vessel is significantly correlated with the occurrence of coronary heart disease (CHD). In this paper, we propose the Residual-block Inspired Coordinate Attention U-Net (RICAU-Net), which incorporates coordinate attention in two distinct manners and a customized combo loss function for lesion-specific coronary artery calcium (CAC) segmentation. This approach aims to tackle the high class-imbalance issue associated with small and sparse lesions, particularly for CAC in the left main coronary artery (LM) which is generally small and the scarcest in the dataset due to its anatomical structure. The proposed method was compared with six different methods using Dice score, precision, and recall. Our approach achieved the highest per-lesion Dice scores for all four lesions, especially for CAC in LM compared to other methods. The ablation studies demonstrated the significance of positional information from the coordinate attention and the customized loss function in segmenting small and sparse lesions with a high class-imbalance problem.
Dealing With Heterogeneous 3D MR Knee Images: A Federated Few-Shot Learning Method With Dual Knowledge Distillation
Apr 18, 2023Abstract:Federated Learning has gained popularity among medical institutions since it enables collaborative training between clients (e.g., hospitals) without aggregating data. However, due to the high cost associated with creating annotations, especially for large 3D image datasets, clinical institutions do not have enough supervised data for training locally. Thus, the performance of the collaborative model is subpar under limited supervision. On the other hand, large institutions have the resources to compile data repositories with high-resolution images and labels. Therefore, individual clients can utilize the knowledge acquired in the public data repositories to mitigate the shortage of private annotated images. In this paper, we propose a federated few-shot learning method with dual knowledge distillation. This method allows joint training with limited annotations across clients without jeopardizing privacy. The supervised learning of the proposed method extracts features from limited labeled data in each client, while the unsupervised data is used to distill both feature and response-based knowledge from a national data repository to further improve the accuracy of the collaborative model and reduce the communication cost. Extensive evaluations are conducted on 3D magnetic resonance knee images from a private clinical dataset. Our proposed method shows superior performance and less training time than other semi-supervised federated learning methods. Codes and additional visualization results are available at https://github.com/hexiaoxiao-cs/fedml-knee.
Autofluorescence Bronchoscopy Video Analysis for Lesion Frame Detection
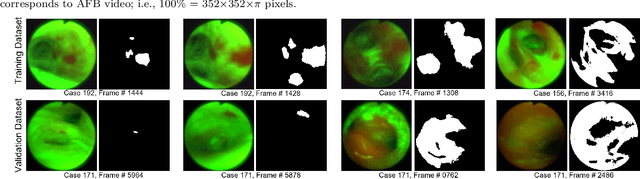
Mar 21, 2023Abstract:Because of the significance of bronchial lesions as indicators of early lung cancer and squamous cell carcinoma, a critical need exists for early detection of bronchial lesions. Autofluorescence bronchoscopy (AFB) is a primary modality used for bronchial lesion detection, as it shows high sensitivity to suspicious lesions. The physician, however, must interactively browse a long video stream to locate lesions, making the search exceedingly tedious and error prone. Unfortunately, limited research has explored the use of automated AFB video analysis for efficient lesion detection. We propose a robust automatic AFB analysis approach that distinguishes informative and uninformative AFB video frames in a video. In addition, for the informative frames, we determine the frames containing potential lesions and delineate candidate lesion regions. Our approach draws upon a combination of computer-based image analysis, machine learning, and deep learning. Thus, the analysis of an AFB video stream becomes more tractable. Tests with patient AFB video indicate that $\ge$97\% of frames were correctly labeled as informative or uninformative. In addition, $\ge$97\% of lesion frames were correctly identified, with false positive and false negative rates $\le$3\%.
Bronchoscopic video synchronization for interactive multimodal inspection of bronchial lesions
Mar 20, 2023



Abstract:With lung cancer being the most fatal cancer worldwide, it is important to detect the disease early. A potentially effective way of detecting early cancer lesions developing along the airway walls (epithelium) is bronchoscopy. To this end, developments in bronchoscopy offer three promising noninvasive modalities for imaging bronchial lesions: white-light bronchoscopy (WLB), autofluorescence bronchoscopy (AFB), and narrow-band imaging (NBI). While these modalities give complementary views of the airway epithelium, the physician must manually inspect each video stream produced by a given modality to locate the suspect cancer lesions. Unfortunately, no effort has been made to rectify this situation by providing efficient quantitative and visual tools for analyzing these video streams. This makes the lesion search process extremely time-consuming and error-prone, thereby making it impractical to utilize these rich data sources effectively. We propose a framework for synchronizing multiple bronchoscopic videos to enable an interactive multimodal analysis of bronchial lesions. Our methods first register the video streams to a reference 3D chest computed-tomography (CT) scan to produce multimodal linkages to the airway tree. Our methods then temporally correlate the videos to one another to enable synchronous visualization of the resulting multimodal data set. Pictorial and quantitative results illustrate the potential of the methods.
ESFPNet: efficient deep learning architecture for real-time lesion segmentation in autofluorescence bronchoscopic video
Jul 15, 2022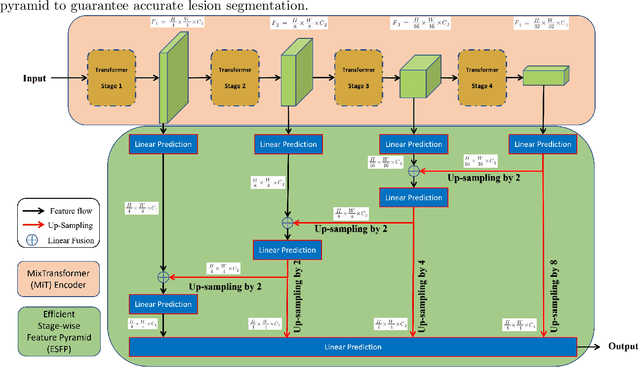

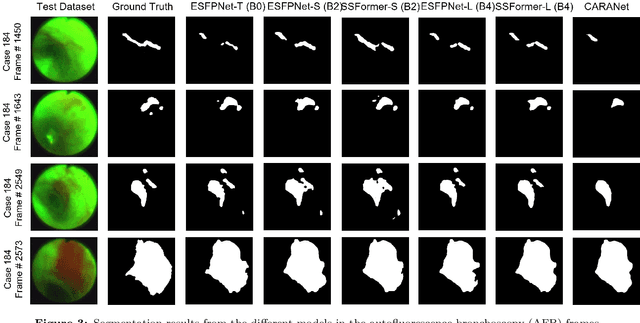
Abstract:Lung cancer tends to be detected at an advanced stage, resulting in a high patient mortality rate. Thus, recent research has focused on early disease detection. Lung cancer generally first appears as lesions developing within the bronchial epithelium of the airway walls. Bronchoscopy is the procedure of choice for effective noninvasive bronchial lesion detection. In particular, autofluorescence bronchoscopy (AFB) discriminates the autofluorescence properties of normal and diseased tissue, whereby lesions appear reddish brown in AFB video frames, while normal tissue appears green. Because recent studies show AFB's ability for high lesion sensitivity, it has become a potentially pivotal method during the standard bronchoscopic airway exam for early-stage lung cancer detection. Unfortunately, manual inspection of AFB video is extremely tedious and error-prone, while limited effort has been expended toward potentially more robust automatic AFB lesion detection and segmentation. We propose a real-time deep learning architecture ESFPNet for robust detection and segmentation of bronchial lesions from an AFB video stream. The architecture features an encoder structure that exploits pretrained Mix Transformer (MiT) encoders and a stage-wise feature pyramid (ESFP) decoder structure. Results from AFB videos derived from lung cancer patient airway exams indicate that our approach gives mean Dice index and IOU values of 0.782 and 0.658, respectively, while having a processing throughput of 27 frames/sec. These values are superior to results achieved by other competing architectures that use Mix transformers or CNN-based encoders. Moreover, the superior performance on the ETIS-LaribPolypDB dataset demonstrates its potential applicability to other domains.
DeepRecon: Joint 2D Cardiac Segmentation and 3D Volume Reconstruction via A Structure-Specific Generative Method
Jun 14, 2022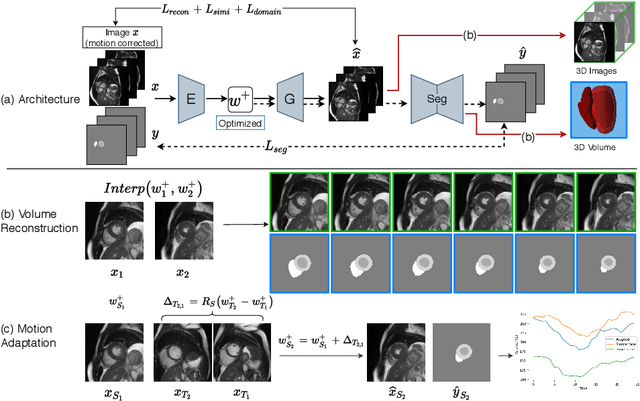
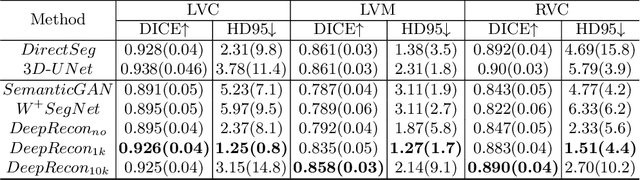
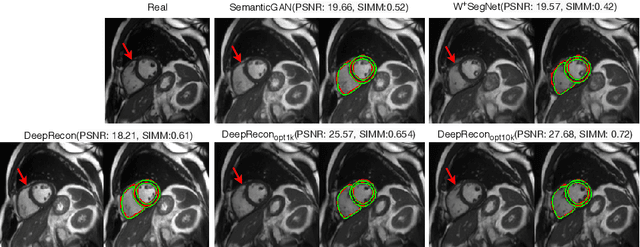
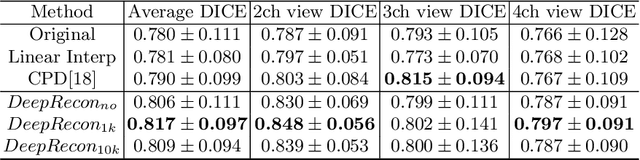
Abstract:Joint 2D cardiac segmentation and 3D volume reconstruction are fundamental to building statistical cardiac anatomy models and understanding functional mechanisms from motion patterns. However, due to the low through-plane resolution of cine MR and high inter-subject variance, accurately segmenting cardiac images and reconstructing the 3D volume are challenging. In this study, we propose an end-to-end latent-space-based framework, DeepRecon, that generates multiple clinically essential outcomes, including accurate image segmentation, synthetic high-resolution 3D image, and 3D reconstructed volume. Our method identifies the optimal latent representation of the cine image that contains accurate semantic information for cardiac structures. In particular, our model jointly generates synthetic images with accurate semantic information and segmentation of the cardiac structures using the optimal latent representation. We further explore downstream applications of 3D shape reconstruction and 4D motion pattern adaptation by the different latent-space manipulation strategies.The simultaneously generated high-resolution images present a high interpretable value to assess the cardiac shape and motion.Experimental results demonstrate the effectiveness of our approach on multiple fronts including 2D segmentation, 3D reconstruction, downstream 4D motion pattern adaption performance.
Modality Bank: Learn multi-modality images across data centers without sharing medical data
Jan 22, 2022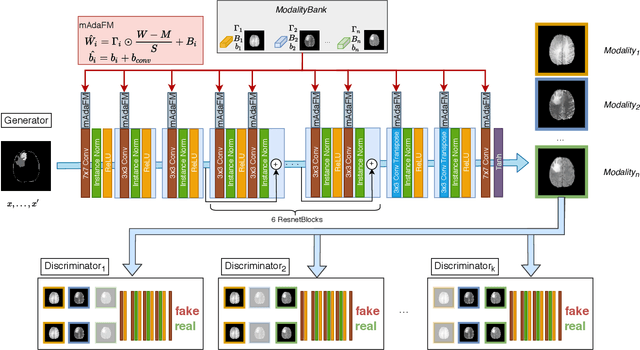
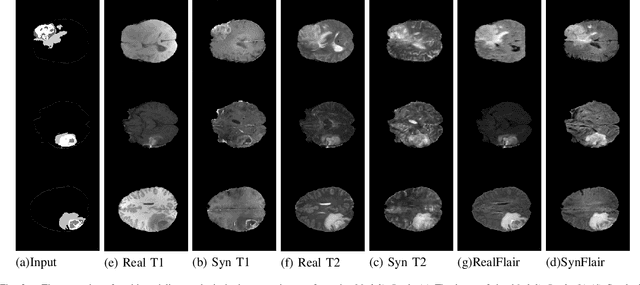
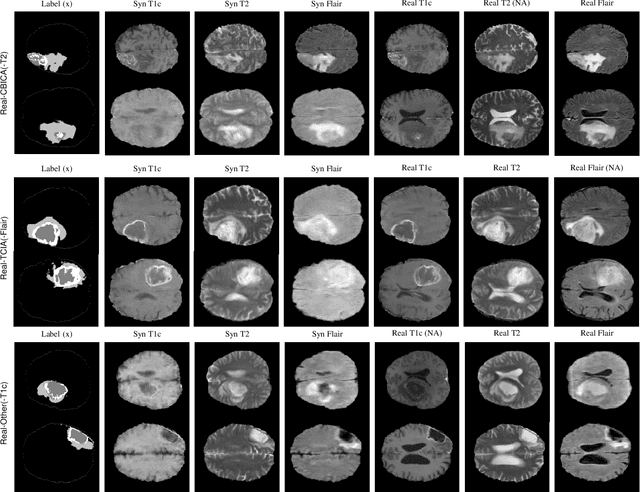
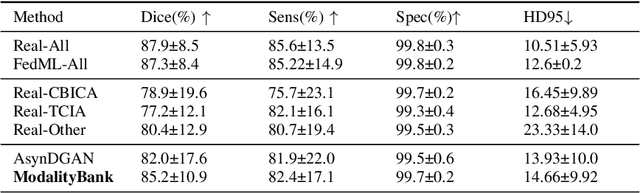
Abstract:Multi-modality images have been widely used and provide comprehensive information for medical image analysis. However, acquiring all modalities among all institutes is costly and often impossible in clinical settings. To leverage more comprehensive multi-modality information, we propose a privacy secured decentralized multi-modality adaptive learning architecture named ModalityBank. Our method could learn a set of effective domain-specific modulation parameters plugged into a common domain-agnostic network. We demonstrate by switching different sets of configurations, the generator could output high-quality images for a specific modality. Our method could also complete the missing modalities across all data centers, thus could be used for modality completion purposes. The downstream task trained from the synthesized multi-modality samples could achieve higher performance than learning from one real data center and achieve close-to-real performance compare with all real images.
DeepTag: An Unsupervised Deep Learning Method for Motion Tracking on Cardiac Tagging Magnetic Resonance Images
Mar 29, 2021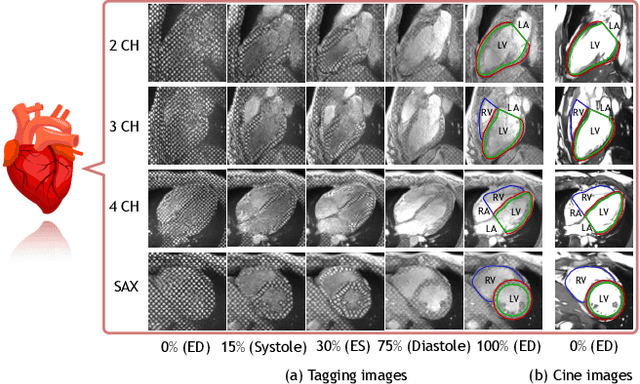
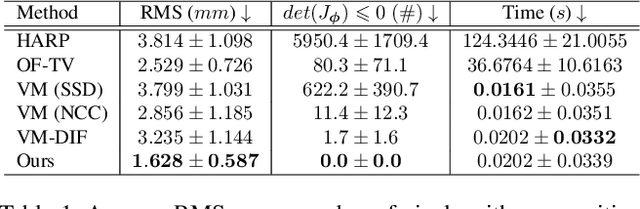
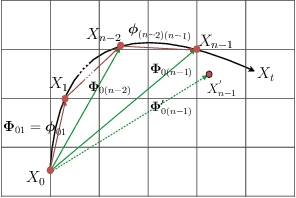
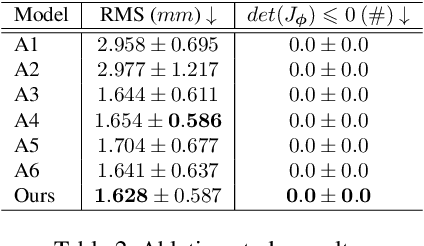
Abstract:Cardiac tagging magnetic resonance imaging (t-MRI) is the gold standard for regional myocardium deformation and cardiac strain estimation. However, this technique has not been widely used in clinical diagnosis, as a result of the difficulty of motion tracking encountered with t-MRI images. In this paper, we propose a novel deep learning-based fully unsupervised method for in vivo motion tracking on t-MRI images. We first estimate the motion field (INF) between any two consecutive t-MRI frames by a bi-directional generative diffeomorphic registration neural network. Using this result, we then estimate the Lagrangian motion field between the reference frame and any other frame through a differentiable composition layer. By utilizing temporal information to perform reasonable estimations on spatio-temporal motion fields, this novel method provides a useful solution for motion tracking and image registration in dynamic medical imaging. Our method has been validated on a representative clinical t-MRI dataset; the experimental results show that our method is superior to conventional motion tracking methods in terms of landmark tracking accuracy and inference efficiency.
Training Federated GANs with Theoretical Guarantees: A Universal Aggregation Approach
Feb 09, 2021



Abstract:Recently, Generative Adversarial Networks (GANs) have demonstrated their potential in federated learning, i.e., learning a centralized model from data privately hosted by multiple sites. A federatedGAN jointly trains a centralized generator and multiple private discriminators hosted at different sites. A major theoretical challenge for the federated GAN is the heterogeneity of the local data distributions. Traditional approaches cannot guarantee to learn the target distribution, which isa mixture of the highly different local distributions. This paper tackles this theoretical challenge, and for the first time, provides a provably correct framework for federated GAN. We propose a new approach called Universal Aggregation, which simulates a centralized discriminator via carefully aggregating the mixture of all private discriminators. We prove that a generator trained with this simulated centralized discriminator can learn the desired target distribution. Through synthetic and real datasets, we show that our method can learn the mixture of largely different distributions where existing federated GAN methods fail.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge