Jennifer Toth
Autofluorescence Bronchoscopy Video Analysis for Lesion Frame Detection
Mar 21, 2023Abstract:Because of the significance of bronchial lesions as indicators of early lung cancer and squamous cell carcinoma, a critical need exists for early detection of bronchial lesions. Autofluorescence bronchoscopy (AFB) is a primary modality used for bronchial lesion detection, as it shows high sensitivity to suspicious lesions. The physician, however, must interactively browse a long video stream to locate lesions, making the search exceedingly tedious and error prone. Unfortunately, limited research has explored the use of automated AFB video analysis for efficient lesion detection. We propose a robust automatic AFB analysis approach that distinguishes informative and uninformative AFB video frames in a video. In addition, for the informative frames, we determine the frames containing potential lesions and delineate candidate lesion regions. Our approach draws upon a combination of computer-based image analysis, machine learning, and deep learning. Thus, the analysis of an AFB video stream becomes more tractable. Tests with patient AFB video indicate that $\ge$97\% of frames were correctly labeled as informative or uninformative. In addition, $\ge$97\% of lesion frames were correctly identified, with false positive and false negative rates $\le$3\%.
Bronchoscopic video synchronization for interactive multimodal inspection of bronchial lesions
Mar 20, 2023



Abstract:With lung cancer being the most fatal cancer worldwide, it is important to detect the disease early. A potentially effective way of detecting early cancer lesions developing along the airway walls (epithelium) is bronchoscopy. To this end, developments in bronchoscopy offer three promising noninvasive modalities for imaging bronchial lesions: white-light bronchoscopy (WLB), autofluorescence bronchoscopy (AFB), and narrow-band imaging (NBI). While these modalities give complementary views of the airway epithelium, the physician must manually inspect each video stream produced by a given modality to locate the suspect cancer lesions. Unfortunately, no effort has been made to rectify this situation by providing efficient quantitative and visual tools for analyzing these video streams. This makes the lesion search process extremely time-consuming and error-prone, thereby making it impractical to utilize these rich data sources effectively. We propose a framework for synchronizing multiple bronchoscopic videos to enable an interactive multimodal analysis of bronchial lesions. Our methods first register the video streams to a reference 3D chest computed-tomography (CT) scan to produce multimodal linkages to the airway tree. Our methods then temporally correlate the videos to one another to enable synchronous visualization of the resulting multimodal data set. Pictorial and quantitative results illustrate the potential of the methods.
ESFPNet: efficient deep learning architecture for real-time lesion segmentation in autofluorescence bronchoscopic video
Jul 15, 2022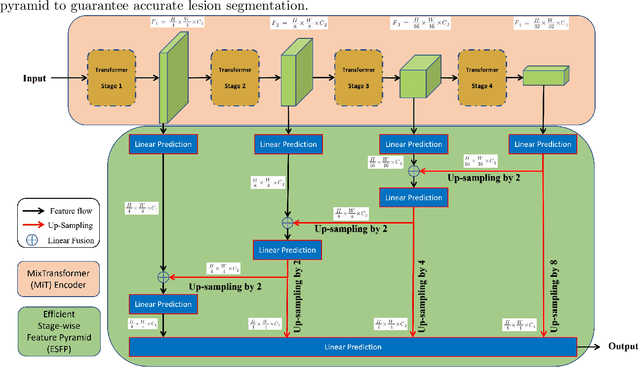

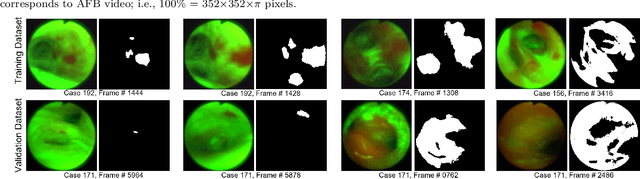
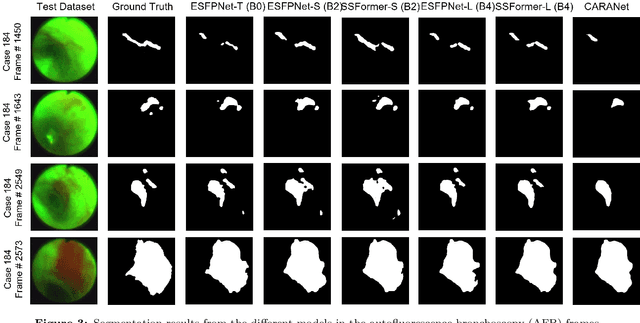
Abstract:Lung cancer tends to be detected at an advanced stage, resulting in a high patient mortality rate. Thus, recent research has focused on early disease detection. Lung cancer generally first appears as lesions developing within the bronchial epithelium of the airway walls. Bronchoscopy is the procedure of choice for effective noninvasive bronchial lesion detection. In particular, autofluorescence bronchoscopy (AFB) discriminates the autofluorescence properties of normal and diseased tissue, whereby lesions appear reddish brown in AFB video frames, while normal tissue appears green. Because recent studies show AFB's ability for high lesion sensitivity, it has become a potentially pivotal method during the standard bronchoscopic airway exam for early-stage lung cancer detection. Unfortunately, manual inspection of AFB video is extremely tedious and error-prone, while limited effort has been expended toward potentially more robust automatic AFB lesion detection and segmentation. We propose a real-time deep learning architecture ESFPNet for robust detection and segmentation of bronchial lesions from an AFB video stream. The architecture features an encoder structure that exploits pretrained Mix Transformer (MiT) encoders and a stage-wise feature pyramid (ESFP) decoder structure. Results from AFB videos derived from lung cancer patient airway exams indicate that our approach gives mean Dice index and IOU values of 0.782 and 0.658, respectively, while having a processing throughput of 27 frames/sec. These values are superior to results achieved by other competing architectures that use Mix transformers or CNN-based encoders. Moreover, the superior performance on the ETIS-LaribPolypDB dataset demonstrates its potential applicability to other domains.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge