Ekaterina Redekop
Computational Mapping of Reactive Stroma in Prostate Cancer Yields Interpretable, Prognostic Biomarkers
Jan 10, 2026Abstract:Current histopathological grading of prostate cancer relies primarily on glandular architecture, largely overlooking the tumor microenvironment. Here, we present PROTAS, a deep learning framework that quantifies reactive stroma (RS) in routine hematoxylin and eosin (H&E) slides and links stromal morphology to underlying biology. PROTAS-defined RS is characterized by nuclear enlargement, collagen disorganization, and transcriptomic enrichment of contractile pathways. PROTAS detects RS robustly in the external Prostate, Lung, Colorectal, and Ovarian (PLCO) dataset and, using domain-adversarial training, generalizes to diagnostic biopsies. In head-to-head comparisons, PROTAS outperforms pathologists for RS detection, and spatial RS features predict biochemical recurrence independently of established prognostic variables (c-index 0.80). By capturing subtle stromal phenotypes associated with tumor progression, PROTAS provides an interpretable, scalable biomarker to refine risk stratification.
CytoFM: The first cytology foundation model
Apr 18, 2025Abstract:Cytology is essential for cancer diagnostics and screening due to its minimally invasive nature. However, the development of robust deep learning models for digital cytology is challenging due to the heterogeneity in staining and preparation methods of samples, differences across organs, and the limited availability of large, diverse, annotated datasets. Developing a task-specific model for every cytology application is impractical and non-cytology-specific foundation models struggle to generalize to tasks in this domain where the emphasis is on cell morphology. To address these challenges, we introduce CytoFM, the first cytology self-supervised foundation model. Using iBOT, a self-supervised Vision Transformer (ViT) training framework incorporating masked image modeling and self-distillation, we pretrain CytoFM on a diverse collection of cytology datasets to learn robust, transferable representations. We evaluate CytoFM on multiple downstream cytology tasks, including breast cancer classification and cell type identification, using an attention-based multiple instance learning framework. Our results demonstrate that CytoFM performs better on two out of three downstream tasks than existing foundation models pretrained on histopathology (UNI) or natural images (iBOT-Imagenet). Visualizations of learned representations demonstrate our model is able to attend to cytologically relevant features. Despite a small pre-training dataset, CytoFM's promising results highlight the ability of task-agnostic pre-training approaches to learn robust and generalizable features from cytology data.
Prototype-Guided Diffusion for Digital Pathology: Achieving Foundation Model Performance with Minimal Clinical Data
Apr 15, 2025
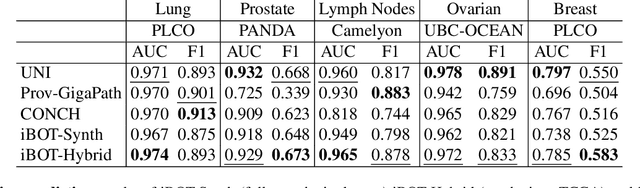
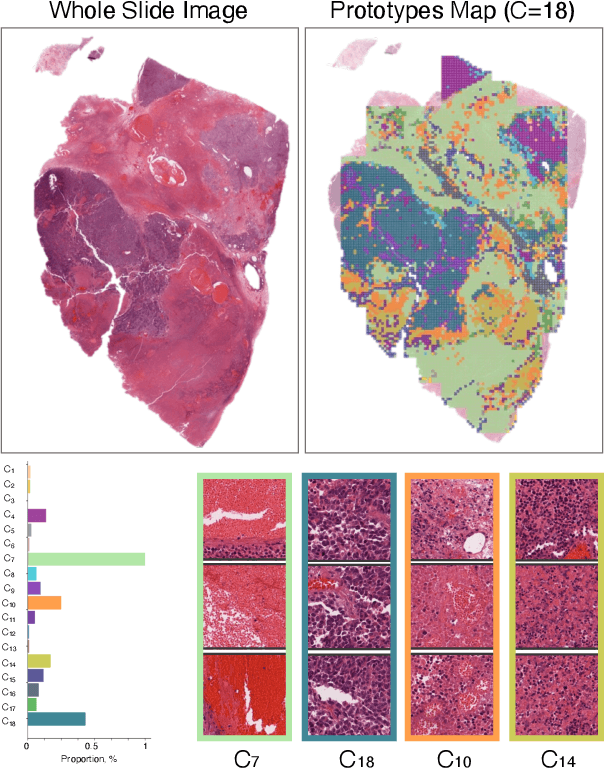
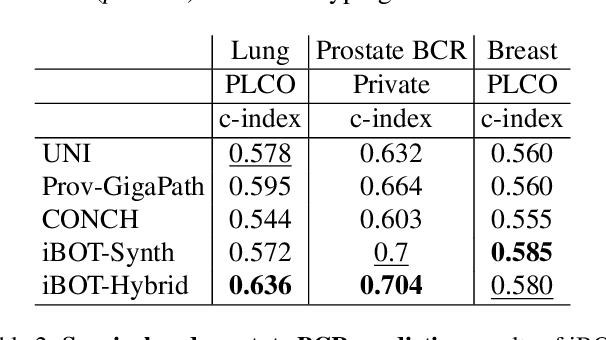
Abstract:Foundation models in digital pathology use massive datasets to learn useful compact feature representations of complex histology images. However, there is limited transparency into what drives the correlation between dataset size and performance, raising the question of whether simply adding more data to increase performance is always necessary. In this study, we propose a prototype-guided diffusion model to generate high-fidelity synthetic pathology data at scale, enabling large-scale self-supervised learning and reducing reliance on real patient samples while preserving downstream performance. Using guidance from histological prototypes during sampling, our approach ensures biologically and diagnostically meaningful variations in the generated data. We demonstrate that self-supervised features trained on our synthetic dataset achieve competitive performance despite using ~60x-760x less data than models trained on large real-world datasets. Notably, models trained using our synthetic data showed statistically comparable or better performance across multiple evaluation metrics and tasks, even when compared to models trained on orders of magnitude larger datasets. Our hybrid approach, combining synthetic and real data, further enhanced performance, achieving top results in several evaluations. These findings underscore the potential of generative AI to create compelling training data for digital pathology, significantly reducing the reliance on extensive clinical datasets and highlighting the efficiency of our approach.
Zero-shot Medical Event Prediction Using a Generative Pre-trained Transformer on Electronic Health Records
Mar 07, 2025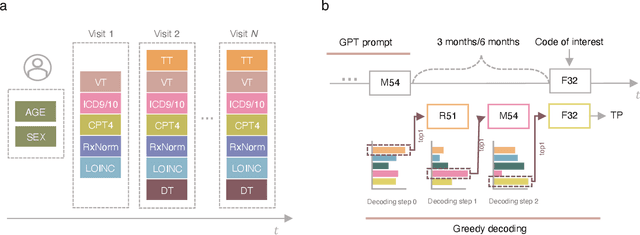
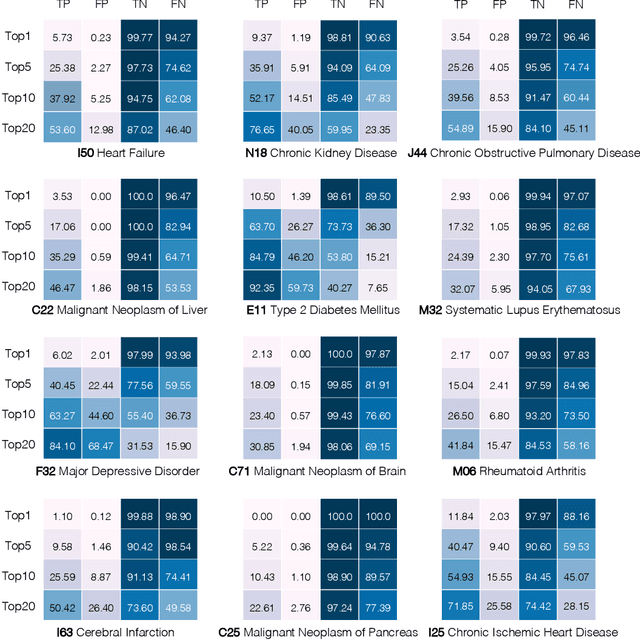
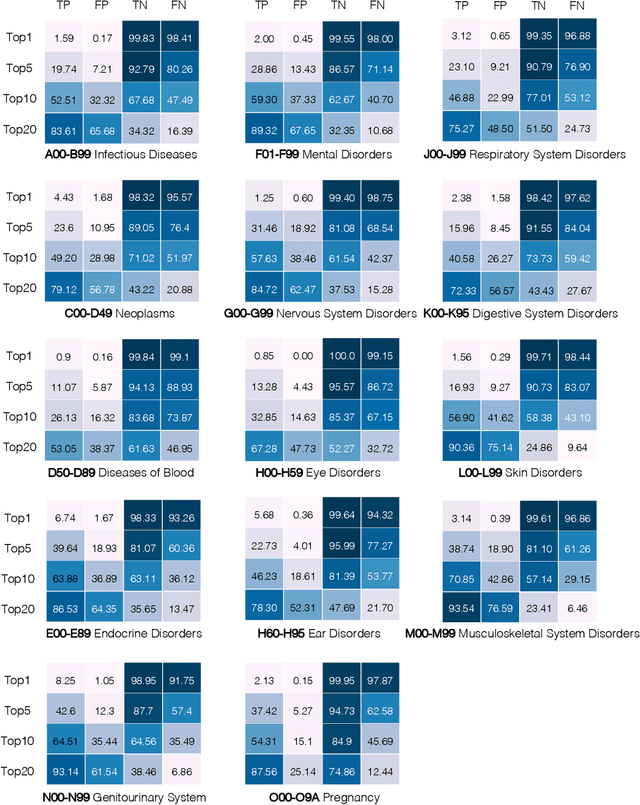
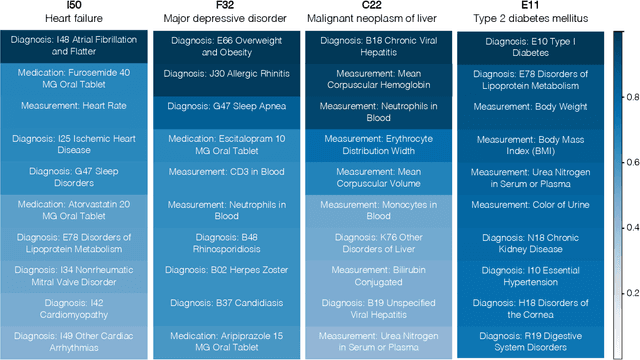
Abstract:Longitudinal data in electronic health records (EHRs) represent an individual`s clinical history through a sequence of codified concepts, including diagnoses, procedures, medications, and laboratory tests. Foundational models, such as generative pre-trained transformers (GPT), can leverage this data to predict future events. While fine-tuning of these models enhances task-specific performance, it is costly, complex, and unsustainable for every target. We show that a foundation model trained on EHRs can perform predictive tasks in a zero-shot manner, eliminating the need for fine-tuning. This study presents the first comprehensive analysis of zero-shot forecasting with GPT-based foundational models in EHRs, introducing a novel pipeline that formulates medical concept prediction as a generative modeling task. Unlike supervised approaches requiring extensive labeled data, our method enables the model to forecast a next medical event purely from a pretraining knowledge. We evaluate performance across multiple time horizons and clinical categories, demonstrating model`s ability to capture latent temporal dependencies and complex patient trajectories without task supervision. Model performance for predicting the next medical concept was evaluated using precision and recall metrics, achieving an average top1 precision of 0.614 and recall of 0.524. For 12 major diagnostic conditions, the model demonstrated strong zero-shot performance, achieving high true positive rates while maintaining low false positives. We demonstrate the power of a foundational EHR GPT model in capturing diverse phenotypes and enabling robust, zero-shot forecasting of clinical outcomes. This capability enhances the versatility of predictive healthcare models and reduces the need for task-specific training, enabling more scalable applications in clinical settings.
Digital Volumetric Biopsy Cores Improve Gleason Grading of Prostate Cancer Using Deep Learning
Sep 12, 2024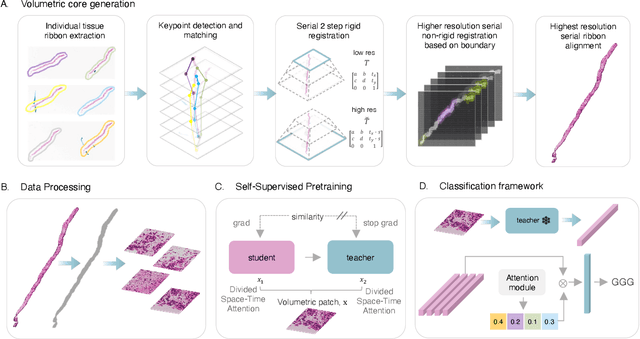
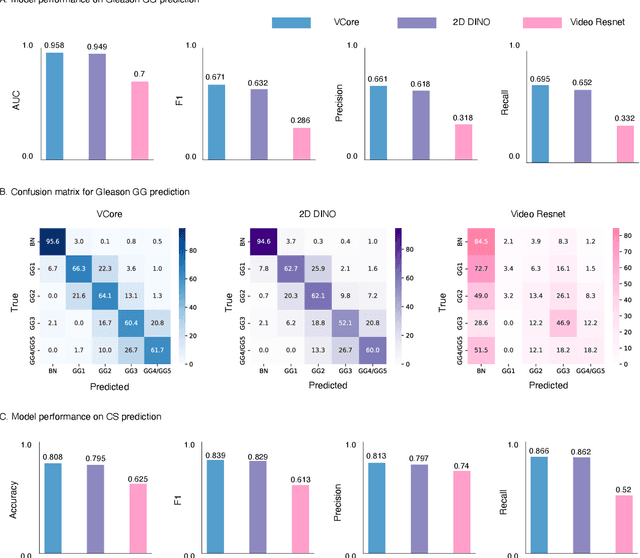

Abstract:Prostate cancer (PCa) was the most frequently diagnosed cancer among American men in 2023. The histological grading of biopsies is essential for diagnosis, and various deep learning-based solutions have been developed to assist with this task. Existing deep learning frameworks are typically applied to individual 2D cross-sections sliced from 3D biopsy tissue specimens. This process impedes the analysis of complex tissue structures such as glands, which can vary depending on the tissue slice examined. We propose a novel digital pathology data source called a "volumetric core," obtained via the extraction and co-alignment of serially sectioned tissue sections using a novel morphology-preserving alignment framework. We trained an attention-based multiple-instance learning (ABMIL) framework on deep features extracted from volumetric patches to automatically classify the Gleason Grade Group (GGG). To handle volumetric patches, we used a modified video transformer with a deep feature extractor pretrained using self-supervised learning. We ran our morphology-preserving alignment framework to construct 10,210 volumetric cores, leaving out 30% for pretraining. The rest of the dataset was used to train ABMIL, which resulted in a 0.958 macro-average AUC, 0.671 F1 score, 0.661 precision, and 0.695 recall averaged across all five GGG significantly outperforming the 2D baselines.
Federated Learning with Research Prototypes for Multi-Center MRI-based Detection of Prostate Cancer with Diverse Histopathology
Jun 11, 2022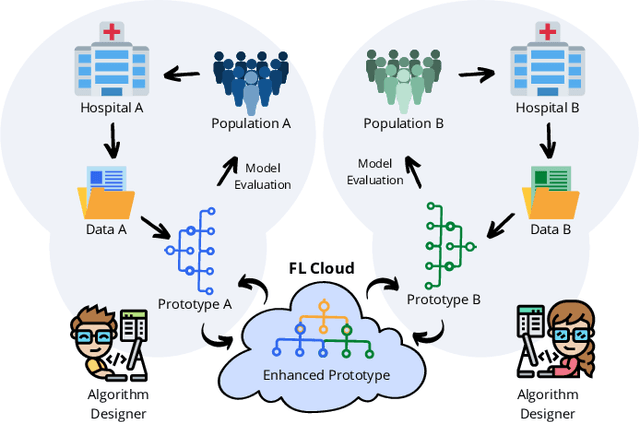
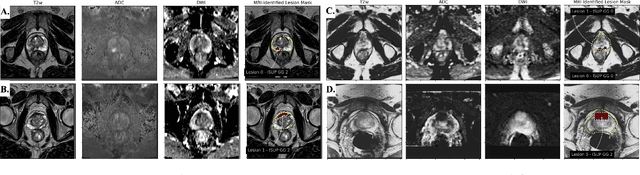
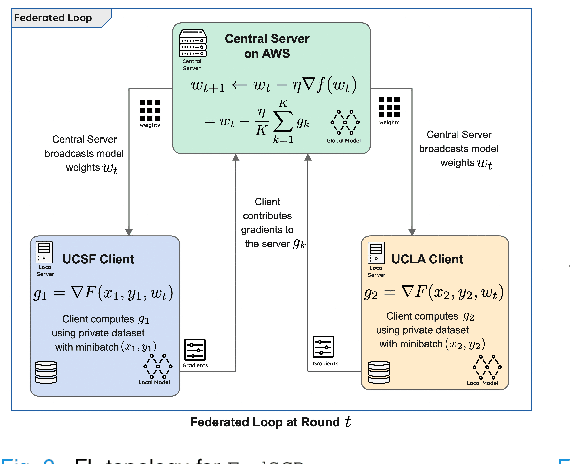
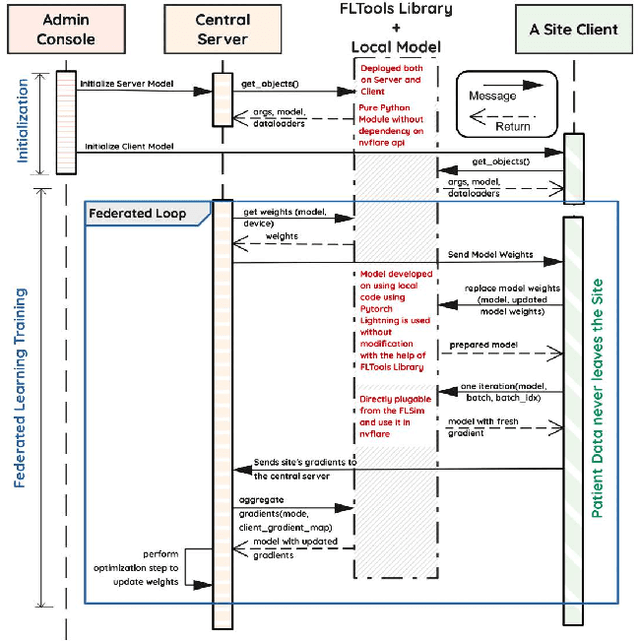
Abstract:Early prostate cancer detection and staging from MRI are extremely challenging tasks for both radiologists and deep learning algorithms, but the potential to learn from large and diverse datasets remains a promising avenue to increase their generalization capability both within- and across clinics. To enable this for prototype-stage algorithms, where the majority of existing research remains, in this paper we introduce a flexible federated learning framework for cross-site training, validation, and evaluation of deep prostate cancer detection algorithms. Our approach utilizes an abstracted representation of the model architecture and data, which allows unpolished prototype deep learning models to be trained without modification using the NVFlare federated learning framework. Our results show increases in prostate cancer detection and classification accuracy using a specialized neural network model and diverse prostate biopsy data collected at two University of California research hospitals, demonstrating the efficacy of our approach in adapting to different datasets and improving MR-biomarker discovery. We open-source our FLtools system, which can be easily adapted to other deep learning projects for medical imaging.
Medical image segmentation with imperfect 3D bounding boxes
Aug 06, 2021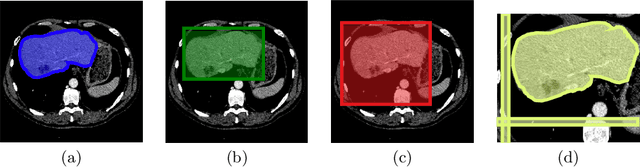



Abstract:The development of high quality medical image segmentation algorithms depends on the availability of large datasets with pixel-level labels. The challenges of collecting such datasets, especially in case of 3D volumes, motivate to develop approaches that can learn from other types of labels that are cheap to obtain, e.g. bounding boxes. We focus on 3D medical images with their corresponding 3D bounding boxes which are considered as series of per-slice non-tight 2D bounding boxes. While current weakly-supervised approaches that use 2D bounding boxes as weak labels can be applied to medical image segmentation, we show that their success is limited in cases when the assumption about the tightness of the bounding boxes breaks. We propose a new bounding box correction framework which is trained on a small set of pixel-level annotations to improve the tightness of a larger set of non-tight bounding box annotations. The effectiveness of our solution is demonstrated by evaluating a known weakly-supervised segmentation approach with and without the proposed bounding box correction algorithm. When the tightness is improved by our solution, the results of the weakly-supervised segmentation become much closer to those of the fully-supervised one.
Uncertainty-based method for improving poorly labeled segmentation datasets
Feb 16, 2021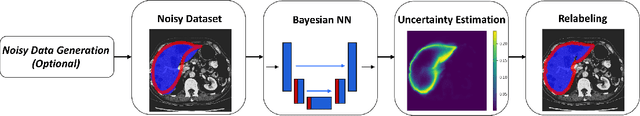
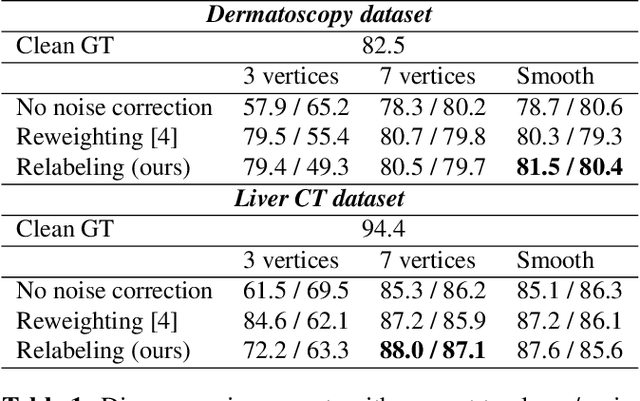
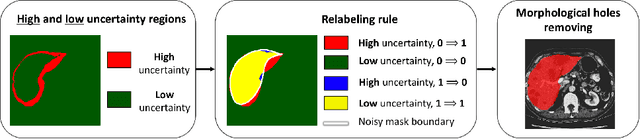
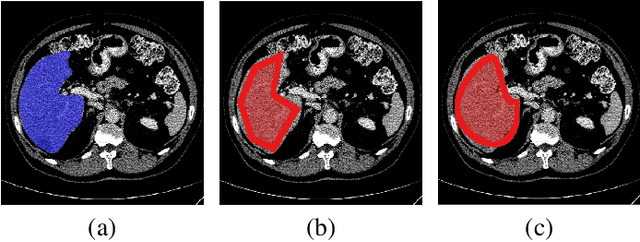
Abstract:The success of modern deep learning algorithms for image segmentation heavily depends on the availability of large datasets with clean pixel-level annotations (masks), where the objects of interest are accurately delineated. Lack of time and expertise during data annotation leads to incorrect boundaries and label noise. It is known that deep convolutional neural networks (DCNNs) can memorize even completely random labels, resulting in poor accuracy. We propose a framework to train binary segmentation DCNNs using sets of unreliable pixel-level annotations. Erroneously labeled pixels are identified based on the estimated aleatoric uncertainty of the segmentation and are relabeled to the true value.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge