Prototype-Guided Diffusion for Digital Pathology: Achieving Foundation Model Performance with Minimal Clinical Data
Paper and Code
Apr 15, 2025
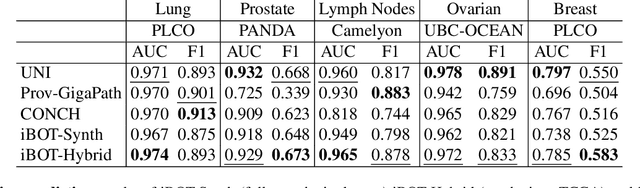
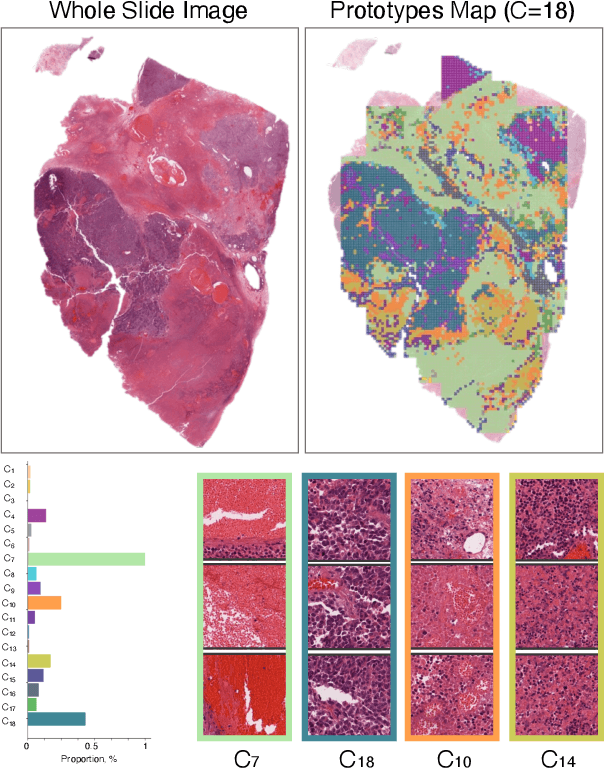
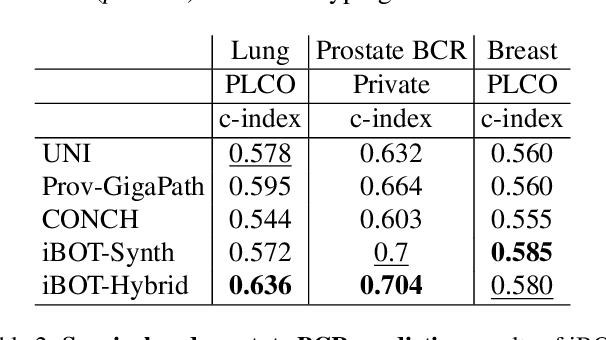
Foundation models in digital pathology use massive datasets to learn useful compact feature representations of complex histology images. However, there is limited transparency into what drives the correlation between dataset size and performance, raising the question of whether simply adding more data to increase performance is always necessary. In this study, we propose a prototype-guided diffusion model to generate high-fidelity synthetic pathology data at scale, enabling large-scale self-supervised learning and reducing reliance on real patient samples while preserving downstream performance. Using guidance from histological prototypes during sampling, our approach ensures biologically and diagnostically meaningful variations in the generated data. We demonstrate that self-supervised features trained on our synthetic dataset achieve competitive performance despite using ~60x-760x less data than models trained on large real-world datasets. Notably, models trained using our synthetic data showed statistically comparable or better performance across multiple evaluation metrics and tasks, even when compared to models trained on orders of magnitude larger datasets. Our hybrid approach, combining synthetic and real data, further enhanced performance, achieving top results in several evaluations. These findings underscore the potential of generative AI to create compelling training data for digital pathology, significantly reducing the reliance on extensive clinical datasets and highlighting the efficiency of our approach.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge