Anthony Sisk
Computational Mapping of Reactive Stroma in Prostate Cancer Yields Interpretable, Prognostic Biomarkers
Jan 10, 2026Abstract:Current histopathological grading of prostate cancer relies primarily on glandular architecture, largely overlooking the tumor microenvironment. Here, we present PROTAS, a deep learning framework that quantifies reactive stroma (RS) in routine hematoxylin and eosin (H&E) slides and links stromal morphology to underlying biology. PROTAS-defined RS is characterized by nuclear enlargement, collagen disorganization, and transcriptomic enrichment of contractile pathways. PROTAS detects RS robustly in the external Prostate, Lung, Colorectal, and Ovarian (PLCO) dataset and, using domain-adversarial training, generalizes to diagnostic biopsies. In head-to-head comparisons, PROTAS outperforms pathologists for RS detection, and spatial RS features predict biochemical recurrence independently of established prognostic variables (c-index 0.80). By capturing subtle stromal phenotypes associated with tumor progression, PROTAS provides an interpretable, scalable biomarker to refine risk stratification.
Prototype-Guided Diffusion for Digital Pathology: Achieving Foundation Model Performance with Minimal Clinical Data
Apr 15, 2025
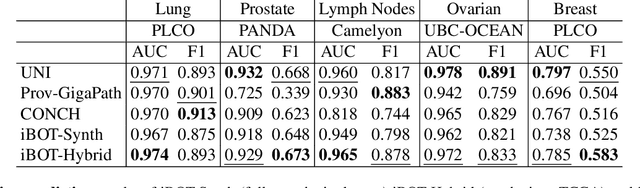
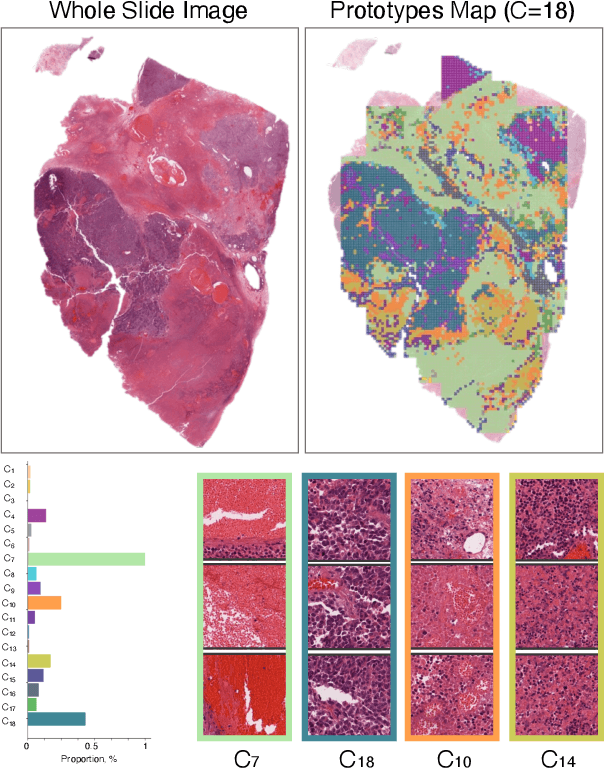
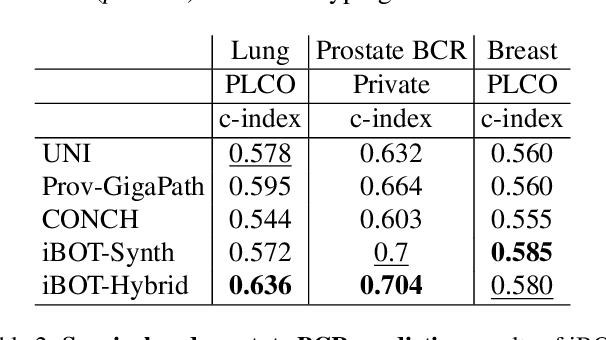
Abstract:Foundation models in digital pathology use massive datasets to learn useful compact feature representations of complex histology images. However, there is limited transparency into what drives the correlation between dataset size and performance, raising the question of whether simply adding more data to increase performance is always necessary. In this study, we propose a prototype-guided diffusion model to generate high-fidelity synthetic pathology data at scale, enabling large-scale self-supervised learning and reducing reliance on real patient samples while preserving downstream performance. Using guidance from histological prototypes during sampling, our approach ensures biologically and diagnostically meaningful variations in the generated data. We demonstrate that self-supervised features trained on our synthetic dataset achieve competitive performance despite using ~60x-760x less data than models trained on large real-world datasets. Notably, models trained using our synthetic data showed statistically comparable or better performance across multiple evaluation metrics and tasks, even when compared to models trained on orders of magnitude larger datasets. Our hybrid approach, combining synthetic and real data, further enhanced performance, achieving top results in several evaluations. These findings underscore the potential of generative AI to create compelling training data for digital pathology, significantly reducing the reliance on extensive clinical datasets and highlighting the efficiency of our approach.
Digital Volumetric Biopsy Cores Improve Gleason Grading of Prostate Cancer Using Deep Learning
Sep 12, 2024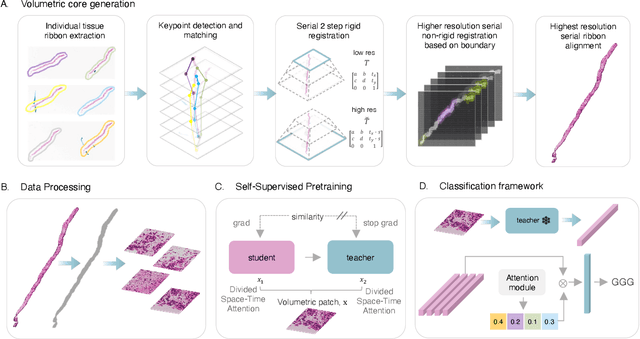
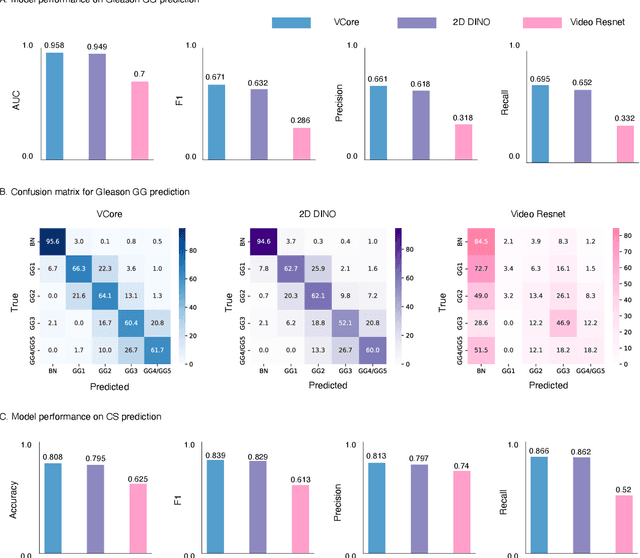

Abstract:Prostate cancer (PCa) was the most frequently diagnosed cancer among American men in 2023. The histological grading of biopsies is essential for diagnosis, and various deep learning-based solutions have been developed to assist with this task. Existing deep learning frameworks are typically applied to individual 2D cross-sections sliced from 3D biopsy tissue specimens. This process impedes the analysis of complex tissue structures such as glands, which can vary depending on the tissue slice examined. We propose a novel digital pathology data source called a "volumetric core," obtained via the extraction and co-alignment of serially sectioned tissue sections using a novel morphology-preserving alignment framework. We trained an attention-based multiple-instance learning (ABMIL) framework on deep features extracted from volumetric patches to automatically classify the Gleason Grade Group (GGG). To handle volumetric patches, we used a modified video transformer with a deep feature extractor pretrained using self-supervised learning. We ran our morphology-preserving alignment framework to construct 10,210 volumetric cores, leaving out 30% for pretraining. The rest of the dataset was used to train ABMIL, which resulted in a 0.958 macro-average AUC, 0.671 F1 score, 0.661 precision, and 0.695 recall averaged across all five GGG significantly outperforming the 2D baselines.
A Multi-resolution Model for Histopathology Image Classification and Localization with Multiple Instance Learning
Nov 05, 2020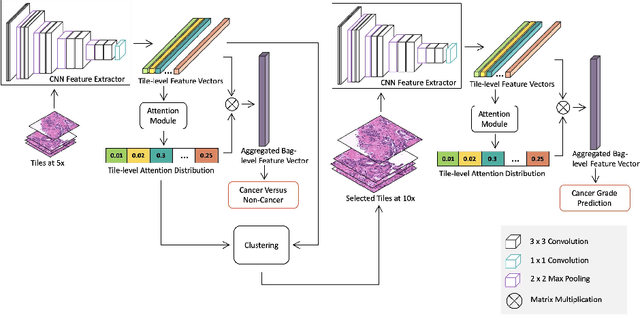
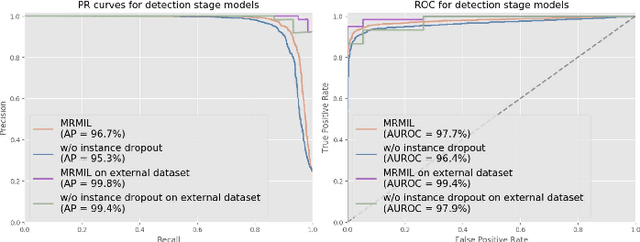
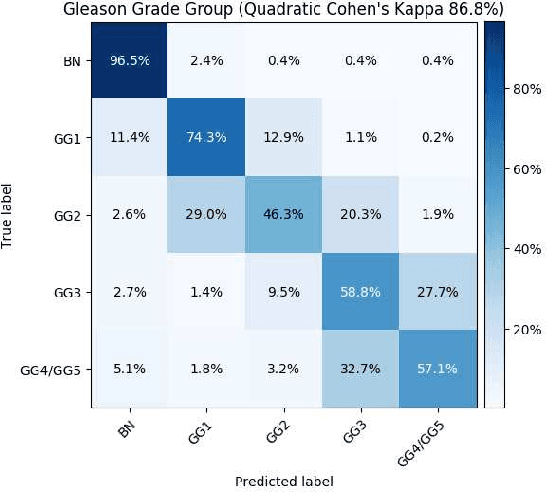
Abstract:Histopathological images provide rich information for disease diagnosis. Large numbers of histopathological images have been digitized into high resolution whole slide images, opening opportunities in developing computational image analysis tools to reduce pathologists' workload and potentially improve inter- and intra- observer agreement. Most previous work on whole slide image analysis has focused on classification or segmentation of small pre-selected regions-of-interest, which requires fine-grained annotation and is non-trivial to extend for large-scale whole slide analysis. In this paper, we proposed a multi-resolution multiple instance learning model that leverages saliency maps to detect suspicious regions for fine-grained grade prediction. Instead of relying on expensive region- or pixel-level annotations, our model can be trained end-to-end with only slide-level labels. The model is developed on a large-scale prostate biopsy dataset containing 20,229 slides from 830 patients. The model achieved 92.7% accuracy, 81.8% Cohen's Kappa for benign, low grade (i.e. Grade group 1) and high grade (i.e. Grade group >= 2) prediction, an area under the receiver operating characteristic curve (AUROC) of 98.2% and an average precision (AP) of 97.4% for differentiating malignant and benign slides. The model obtained an AUROC of 99.4% and an AP of 99.8% for cancer detection on an external dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge