William Speier
Computational Mapping of Reactive Stroma in Prostate Cancer Yields Interpretable, Prognostic Biomarkers
Jan 10, 2026Abstract:Current histopathological grading of prostate cancer relies primarily on glandular architecture, largely overlooking the tumor microenvironment. Here, we present PROTAS, a deep learning framework that quantifies reactive stroma (RS) in routine hematoxylin and eosin (H&E) slides and links stromal morphology to underlying biology. PROTAS-defined RS is characterized by nuclear enlargement, collagen disorganization, and transcriptomic enrichment of contractile pathways. PROTAS detects RS robustly in the external Prostate, Lung, Colorectal, and Ovarian (PLCO) dataset and, using domain-adversarial training, generalizes to diagnostic biopsies. In head-to-head comparisons, PROTAS outperforms pathologists for RS detection, and spatial RS features predict biochemical recurrence independently of established prognostic variables (c-index 0.80). By capturing subtle stromal phenotypes associated with tumor progression, PROTAS provides an interpretable, scalable biomarker to refine risk stratification.
Atherosclerosis through Hierarchical Explainable Neural Network Analysis
Jul 10, 2025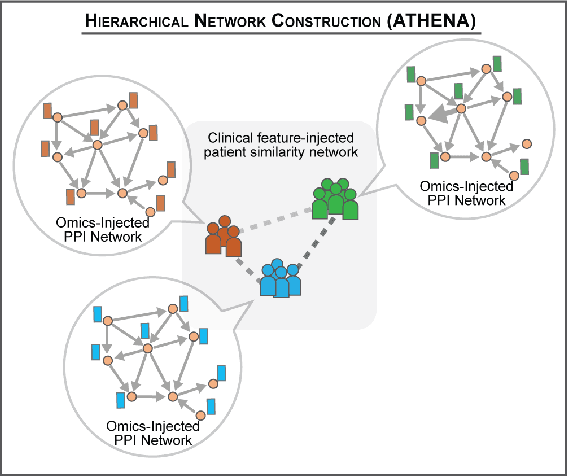
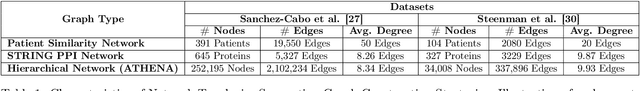
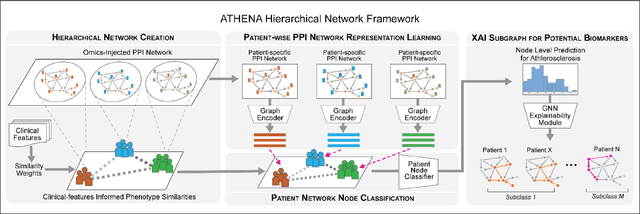
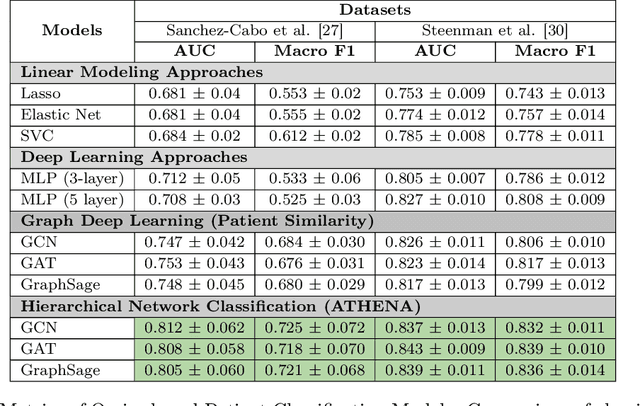
Abstract:In this work, we study the problem pertaining to personalized classification of subclinical atherosclerosis by developing a hierarchical graph neural network framework to leverage two characteristic modalities of a patient: clinical features within the context of the cohort, and molecular data unique to individual patients. Current graph-based methods for disease classification detect patient-specific molecular fingerprints, but lack consistency and comprehension regarding cohort-wide features, which are an essential requirement for understanding pathogenic phenotypes across diverse atherosclerotic trajectories. Furthermore, understanding patient subtypes often considers clinical feature similarity in isolation, without integration of shared pathogenic interdependencies among patients. To address these challenges, we introduce ATHENA: Atherosclerosis Through Hierarchical Explainable Neural Network Analysis, which constructs a novel hierarchical network representation through integrated modality learning; subsequently, it optimizes learned patient-specific molecular fingerprints that reflect individual omics data, enforcing consistency with cohort-wide patterns. With a primary clinical dataset of 391 patients, we demonstrate that this heterogeneous alignment of clinical features with molecular interaction patterns has significantly boosted subclinical atherosclerosis classification performance across various baselines by up to 13% in area under the receiver operating curve (AUC) and 20% in F1 score. Taken together, ATHENA enables mechanistically-informed patient subtype discovery through explainable AI (XAI)-driven subnetwork clustering; this novel integration framework strengthens personalized intervention strategies, thereby improving the prediction of atherosclerotic disease progression and management of their clinical actionable outcomes.
CytoFM: The first cytology foundation model
Apr 18, 2025Abstract:Cytology is essential for cancer diagnostics and screening due to its minimally invasive nature. However, the development of robust deep learning models for digital cytology is challenging due to the heterogeneity in staining and preparation methods of samples, differences across organs, and the limited availability of large, diverse, annotated datasets. Developing a task-specific model for every cytology application is impractical and non-cytology-specific foundation models struggle to generalize to tasks in this domain where the emphasis is on cell morphology. To address these challenges, we introduce CytoFM, the first cytology self-supervised foundation model. Using iBOT, a self-supervised Vision Transformer (ViT) training framework incorporating masked image modeling and self-distillation, we pretrain CytoFM on a diverse collection of cytology datasets to learn robust, transferable representations. We evaluate CytoFM on multiple downstream cytology tasks, including breast cancer classification and cell type identification, using an attention-based multiple instance learning framework. Our results demonstrate that CytoFM performs better on two out of three downstream tasks than existing foundation models pretrained on histopathology (UNI) or natural images (iBOT-Imagenet). Visualizations of learned representations demonstrate our model is able to attend to cytologically relevant features. Despite a small pre-training dataset, CytoFM's promising results highlight the ability of task-agnostic pre-training approaches to learn robust and generalizable features from cytology data.
Prototype-Guided Diffusion for Digital Pathology: Achieving Foundation Model Performance with Minimal Clinical Data
Apr 15, 2025
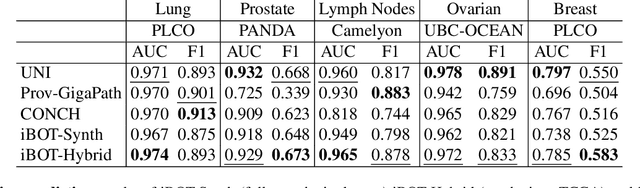
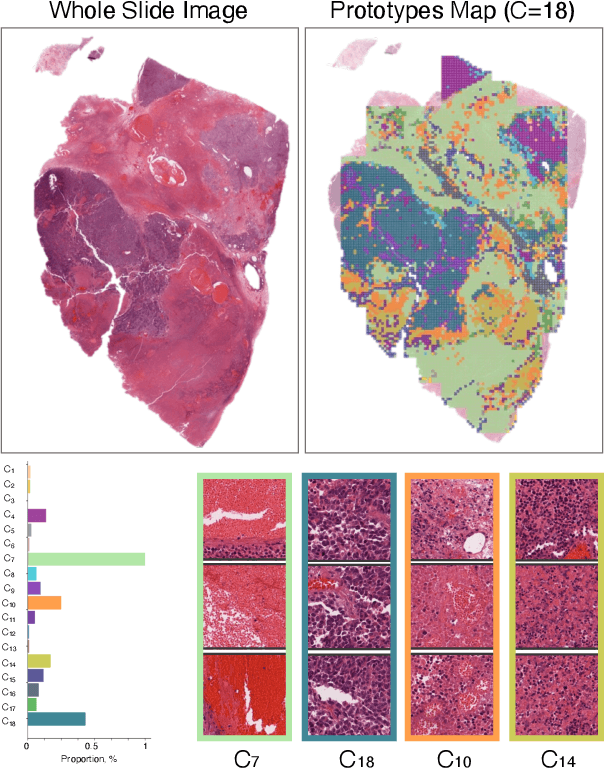
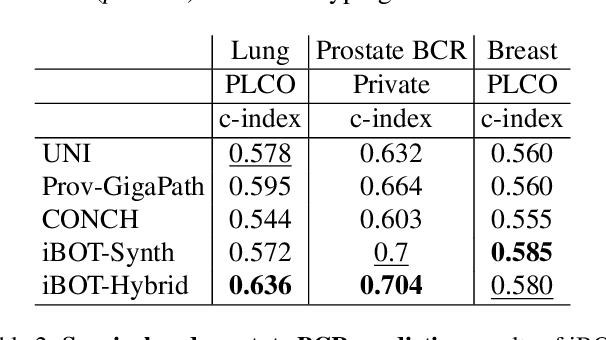
Abstract:Foundation models in digital pathology use massive datasets to learn useful compact feature representations of complex histology images. However, there is limited transparency into what drives the correlation between dataset size and performance, raising the question of whether simply adding more data to increase performance is always necessary. In this study, we propose a prototype-guided diffusion model to generate high-fidelity synthetic pathology data at scale, enabling large-scale self-supervised learning and reducing reliance on real patient samples while preserving downstream performance. Using guidance from histological prototypes during sampling, our approach ensures biologically and diagnostically meaningful variations in the generated data. We demonstrate that self-supervised features trained on our synthetic dataset achieve competitive performance despite using ~60x-760x less data than models trained on large real-world datasets. Notably, models trained using our synthetic data showed statistically comparable or better performance across multiple evaluation metrics and tasks, even when compared to models trained on orders of magnitude larger datasets. Our hybrid approach, combining synthetic and real data, further enhanced performance, achieving top results in several evaluations. These findings underscore the potential of generative AI to create compelling training data for digital pathology, significantly reducing the reliance on extensive clinical datasets and highlighting the efficiency of our approach.
Zero-shot Medical Event Prediction Using a Generative Pre-trained Transformer on Electronic Health Records
Mar 07, 2025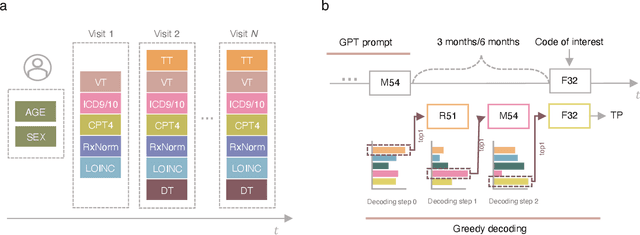
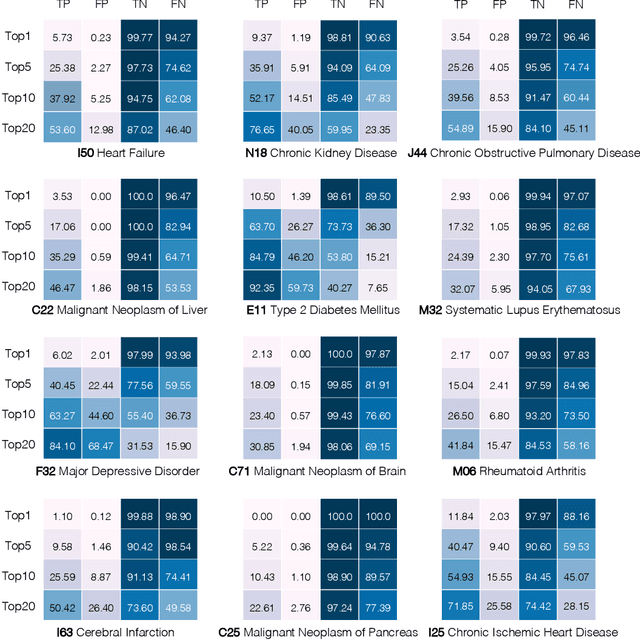
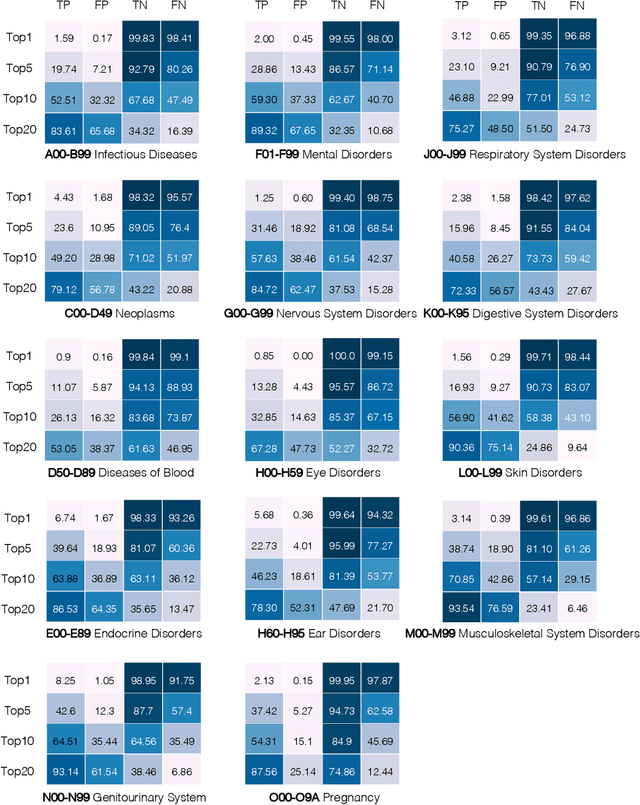
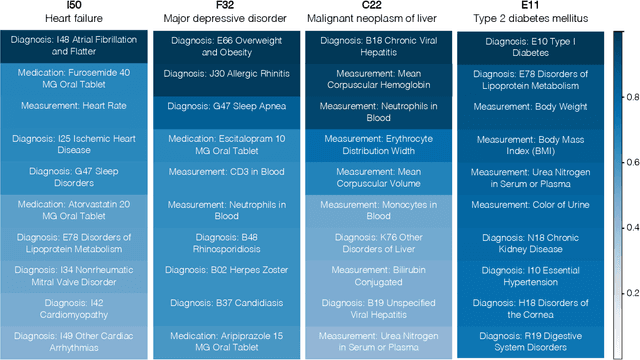
Abstract:Longitudinal data in electronic health records (EHRs) represent an individual`s clinical history through a sequence of codified concepts, including diagnoses, procedures, medications, and laboratory tests. Foundational models, such as generative pre-trained transformers (GPT), can leverage this data to predict future events. While fine-tuning of these models enhances task-specific performance, it is costly, complex, and unsustainable for every target. We show that a foundation model trained on EHRs can perform predictive tasks in a zero-shot manner, eliminating the need for fine-tuning. This study presents the first comprehensive analysis of zero-shot forecasting with GPT-based foundational models in EHRs, introducing a novel pipeline that formulates medical concept prediction as a generative modeling task. Unlike supervised approaches requiring extensive labeled data, our method enables the model to forecast a next medical event purely from a pretraining knowledge. We evaluate performance across multiple time horizons and clinical categories, demonstrating model`s ability to capture latent temporal dependencies and complex patient trajectories without task supervision. Model performance for predicting the next medical concept was evaluated using precision and recall metrics, achieving an average top1 precision of 0.614 and recall of 0.524. For 12 major diagnostic conditions, the model demonstrated strong zero-shot performance, achieving high true positive rates while maintaining low false positives. We demonstrate the power of a foundational EHR GPT model in capturing diverse phenotypes and enabling robust, zero-shot forecasting of clinical outcomes. This capability enhances the versatility of predictive healthcare models and reduces the need for task-specific training, enabling more scalable applications in clinical settings.
Evaluation Of P300 Speller Performance Using Large Language Models Along With Cross-Subject Training
Oct 19, 2024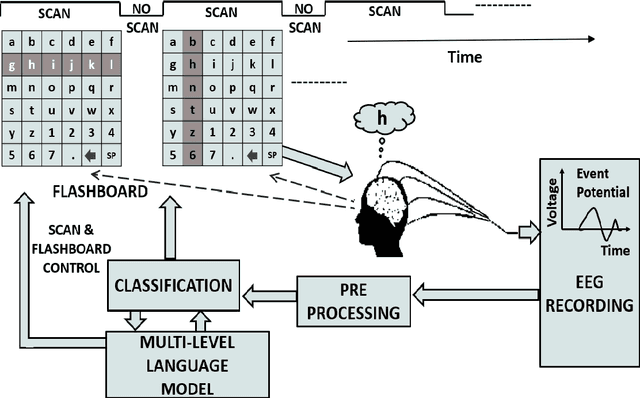
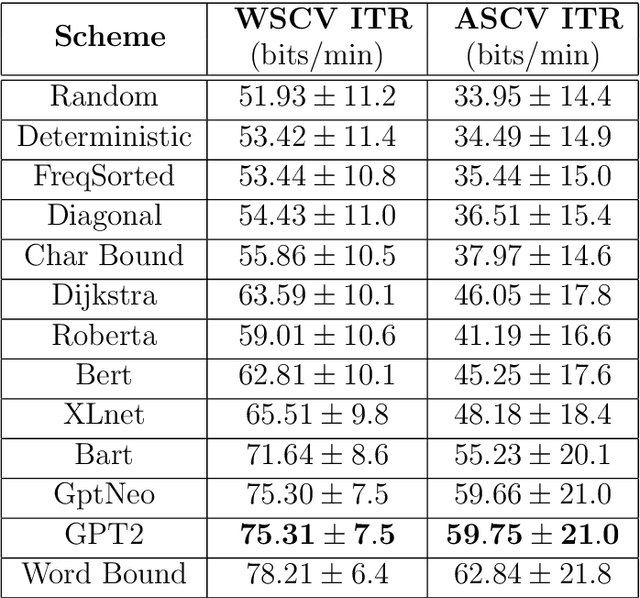
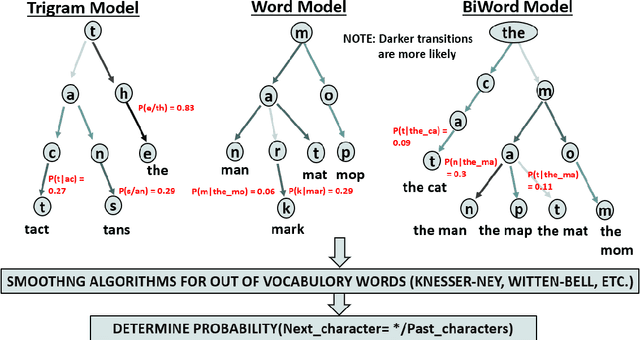

Abstract:Amyotrophic lateral sclerosis (ALS), a progressive neuromuscular degenerative disease, severely restricts patient communication capacity within a few years of onset, resulting in a significant deterioration of quality of life. The P300 speller brain computer interface (BCI) offers an alternative communication medium by leveraging a subject's EEG response to characters traditionally highlighted on a character grid on a graphical user interface (GUI). A recurring theme in P300-based research is enhancing performance to enable faster subject interaction. This study builds on that theme by addressing key limitations, particularly in the training of multi-subject classifiers, and by integrating advanced language models to optimize stimuli presentation and word prediction, thereby improving communication efficiency. Furthermore, various advanced large language models such as Generative Pre-Trained Transformer (GPT2), BERT, and BART, alongside Dijkstra's algorithm, are utilized to optimize stimuli and provide word completion choices based on the spelling history. In addition, a multi-layered smoothing approach is applied to allow for out-of-vocabulary (OOV) words. By conducting extensive simulations based on randomly sampled EEG data from subjects, we show substantial speed improvements in typing passages that include rare and out-of-vocabulary (OOV) words, with the extent of improvement varying depending on the language model utilized. The gains through such character-level interface optimizations are approximately 10%, and GPT2 for multi-word prediction provides gains of around 40%. In particular, some large language models achieve performance levels within 10% of the theoretical performance limits established in this study. In addition, both within and across subjects, training techniques are explored, and speed improvements are shown to hold in both cases.
Reducing Overtreatment of Indeterminate Thyroid Nodules Using a Multimodal Deep Learning Model
Sep 27, 2024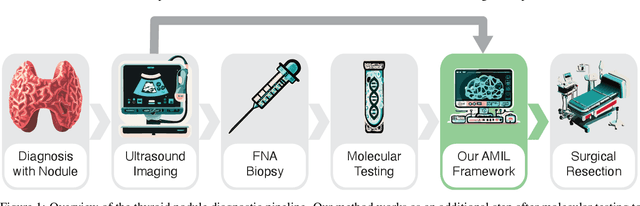
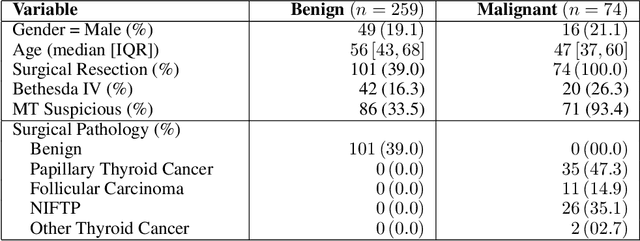
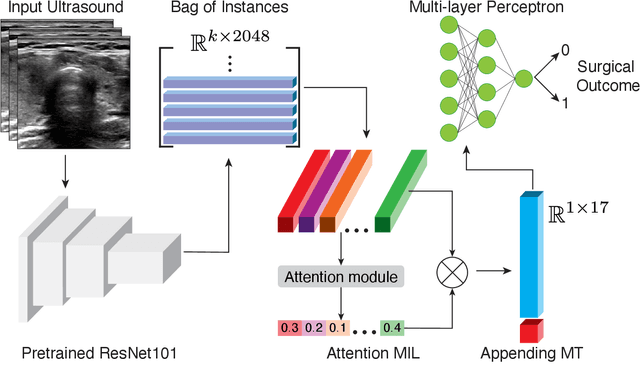
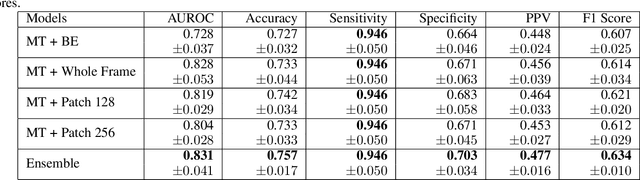
Abstract:Objective: Molecular testing (MT) classifies cytologically indeterminate thyroid nodules as benign or malignant with high sensitivity but low positive predictive value (PPV), only using molecular profiles, ignoring ultrasound (US) imaging and biopsy. We address this limitation by applying attention multiple instance learning (AMIL) to US images. Methods: We retrospectively reviewed 333 patients with indeterminate thyroid nodules at UCLA medical center (259 benign, 74 malignant). A multi-modal deep learning AMIL model was developed, combining US images and MT to classify the nodules as benign or malignant and enhance the malignancy risk stratification of MT. Results: The final AMIL model matched MT sensitivity (0.946) while significantly improving PPV (0.477 vs 0.448 for MT alone), indicating fewer false positives while maintaining high sensitivity. Conclusion: Our approach reduces false positives compared to MT while maintaining the same ability to identify positive cases, potentially reducing unnecessary benign thyroid resections in patients with indeterminate nodules.
Digital Volumetric Biopsy Cores Improve Gleason Grading of Prostate Cancer Using Deep Learning
Sep 12, 2024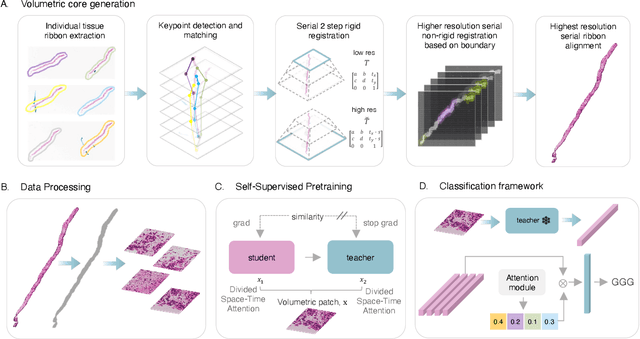
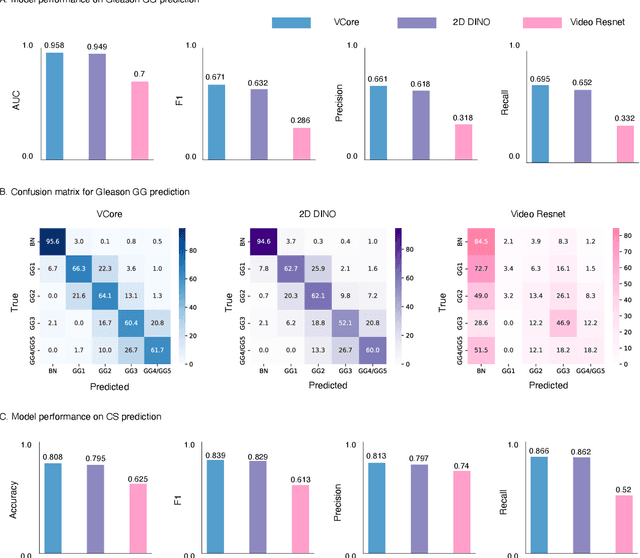
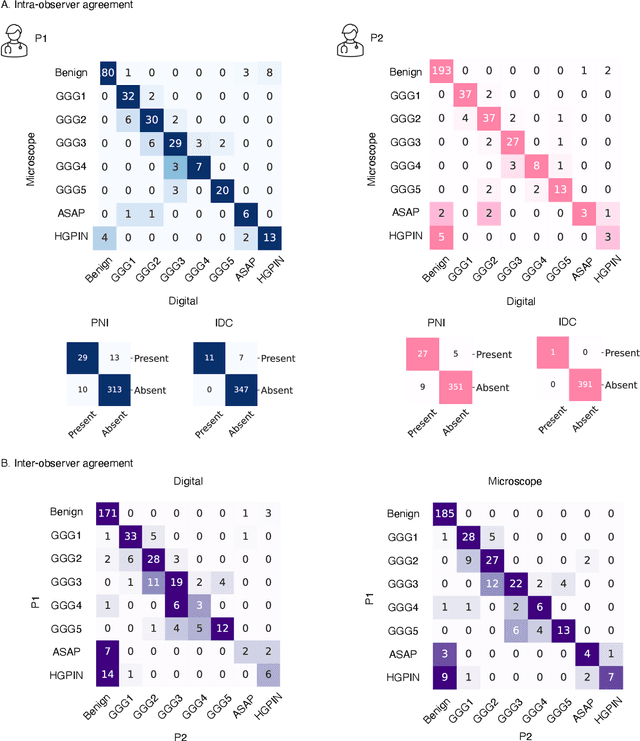

Abstract:Prostate cancer (PCa) was the most frequently diagnosed cancer among American men in 2023. The histological grading of biopsies is essential for diagnosis, and various deep learning-based solutions have been developed to assist with this task. Existing deep learning frameworks are typically applied to individual 2D cross-sections sliced from 3D biopsy tissue specimens. This process impedes the analysis of complex tissue structures such as glands, which can vary depending on the tissue slice examined. We propose a novel digital pathology data source called a "volumetric core," obtained via the extraction and co-alignment of serially sectioned tissue sections using a novel morphology-preserving alignment framework. We trained an attention-based multiple-instance learning (ABMIL) framework on deep features extracted from volumetric patches to automatically classify the Gleason Grade Group (GGG). To handle volumetric patches, we used a modified video transformer with a deep feature extractor pretrained using self-supervised learning. We ran our morphology-preserving alignment framework to construct 10,210 volumetric cores, leaving out 30% for pretraining. The rest of the dataset was used to train ABMIL, which resulted in a 0.958 macro-average AUC, 0.671 F1 score, 0.661 precision, and 0.695 recall averaged across all five GGG significantly outperforming the 2D baselines.
High Performance P300 Spellers Using GPT2 Word Prediction With Cross-Subject Training
May 22, 2024Abstract:Amyotrophic lateral sclerosis (ALS) severely impairs patients' ability to communicate, often leading to a decline in their quality of life within a few years of diagnosis. The P300 speller brain-computer interface (BCI) offers an alternative communication method by interpreting a subject's EEG response to characters presented on a grid interface. This paper addresses the common speed limitations encountered in training efficient P300-based multi-subject classifiers by introducing innovative "across-subject" classifiers. We leverage a combination of the second-generation Generative Pre-Trained Transformer (GPT2) and Dijkstra's algorithm to optimize stimuli and suggest word completion choices based on typing history. Additionally, we employ a multi-layered smoothing technique to accommodate out-of-vocabulary (OOV) words. Through extensive simulations involving random sampling of EEG data from subjects, we demonstrate significant speed enhancements in typing passages containing rare and OOV words. These optimizations result in approximately 10% improvement in character-level typing speed and up to 40% improvement in multi-word prediction. We demonstrate that augmenting standard row/column highlighting techniques with layered word prediction yields close-to-optimal performance. Furthermore, we explore both "within-subject" and "across-subject" training techniques, showing that speed improvements are consistent across both approaches.
Ultrasound Image Enhancement using CycleGAN and Perceptual Loss
Dec 18, 2023Abstract:Purpose: The objective of this work is to introduce an advanced framework designed to enhance ultrasound images, especially those captured by portable hand-held devices, which often produce lower quality images due to hardware constraints. Additionally, this framework is uniquely capable of effectively handling non-registered input ultrasound image pairs, addressing a common challenge in medical imaging. Materials and Methods: In this retrospective study, we utilized an enhanced generative adversarial network (CycleGAN) model for ultrasound image enhancement across five organ systems. Perceptual loss, derived from deep features of pretrained neural networks, is applied to ensure the human-perceptual quality of the enhanced images. These images are compared with paired images acquired from high resolution devices to demonstrate the model's ability to generate realistic high-quality images across organ systems. Results: Preliminary validation of the framework reveals promising performance metrics. The model generates images that result in a Structural Similarity Index (SSI) score of 0.722, Locally Normalized Cross-Correlation (LNCC) score of 0.902 and 28.802 for the Peak Signal-to-Noise Ratio (PSNR) metric. Conclusion: This work presents a significant advancement in medical imaging through the development of a CycleGAN model enhanced with Perceptual Loss (PL), effectively bridging the quality gap between ultrasound images from varied devices. By training on paired images, the model not only improves image quality but also ensures the preservation of vital anatomic structural content. This approach may improve equity in access to healthcare by enhancing portable device capabilities, although further validation and optimizations are necessary for broader clinical application.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge