Melika Emami
Zero-shot Medical Event Prediction Using a Generative Pre-trained Transformer on Electronic Health Records
Mar 07, 2025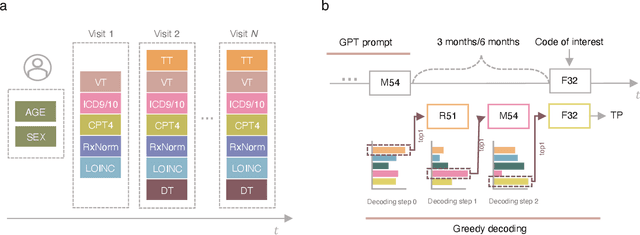
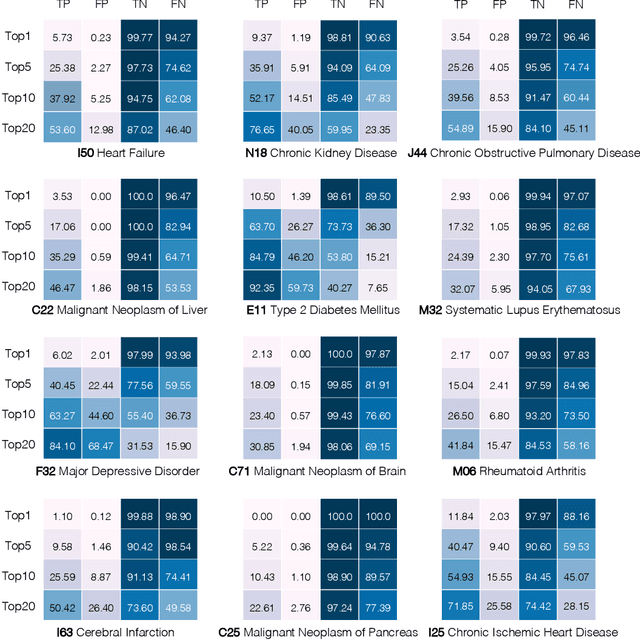
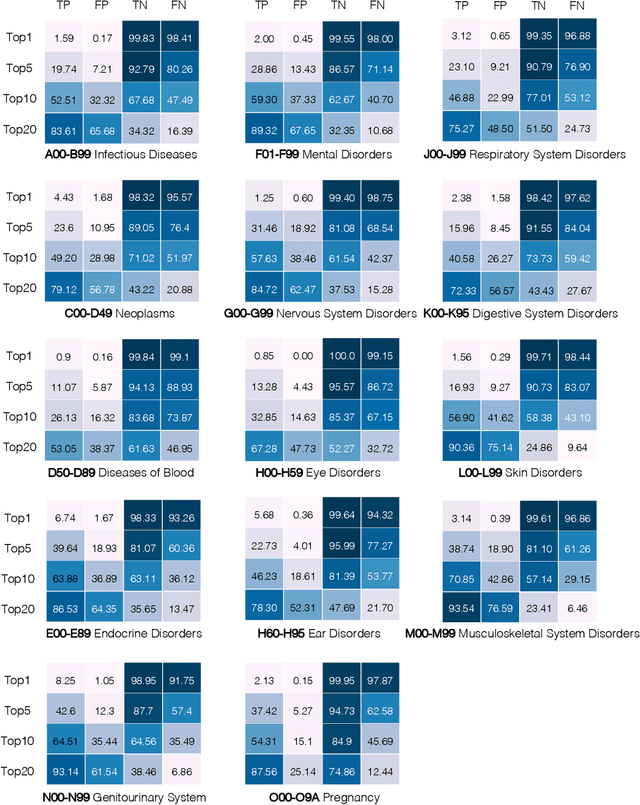
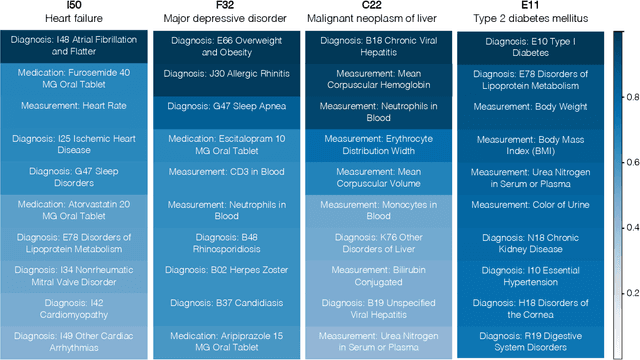
Abstract:Longitudinal data in electronic health records (EHRs) represent an individual`s clinical history through a sequence of codified concepts, including diagnoses, procedures, medications, and laboratory tests. Foundational models, such as generative pre-trained transformers (GPT), can leverage this data to predict future events. While fine-tuning of these models enhances task-specific performance, it is costly, complex, and unsustainable for every target. We show that a foundation model trained on EHRs can perform predictive tasks in a zero-shot manner, eliminating the need for fine-tuning. This study presents the first comprehensive analysis of zero-shot forecasting with GPT-based foundational models in EHRs, introducing a novel pipeline that formulates medical concept prediction as a generative modeling task. Unlike supervised approaches requiring extensive labeled data, our method enables the model to forecast a next medical event purely from a pretraining knowledge. We evaluate performance across multiple time horizons and clinical categories, demonstrating model`s ability to capture latent temporal dependencies and complex patient trajectories without task supervision. Model performance for predicting the next medical concept was evaluated using precision and recall metrics, achieving an average top1 precision of 0.614 and recall of 0.524. For 12 major diagnostic conditions, the model demonstrated strong zero-shot performance, achieving high true positive rates while maintaining low false positives. We demonstrate the power of a foundational EHR GPT model in capturing diverse phenotypes and enabling robust, zero-shot forecasting of clinical outcomes. This capability enhances the versatility of predictive healthcare models and reduces the need for task-specific training, enabling more scalable applications in clinical settings.
LLMs in Biomedicine: A study on clinical Named Entity Recognition
Apr 10, 2024



Abstract:Large Language Models (LLMs) demonstrate remarkable versatility in various NLP tasks but encounter distinct challenges in biomedicine due to medical language complexities and data scarcity. This paper investigates the application of LLMs in the medical domain by exploring strategies to enhance their performance for the Named-Entity Recognition (NER) task. Specifically, our study reveals the importance of meticulously designed prompts in biomedicine. Strategic selection of in-context examples yields a notable improvement, showcasing ~15-20\% increase in F1 score across all benchmark datasets for few-shot clinical NER. Additionally, our findings suggest that integrating external resources through prompting strategies can bridge the gap between general-purpose LLM proficiency and the specialized demands of medical NER. Leveraging a medical knowledge base, our proposed method inspired by Retrieval-Augmented Generation (RAG) can boost the F1 score of LLMs for zero-shot clinical NER. We will release the code upon publication.
Estimation of embedding vectors in high dimensions
Dec 12, 2023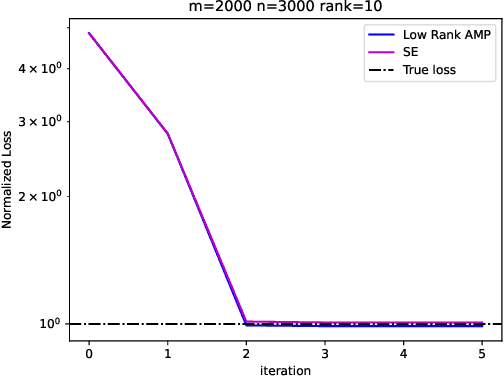
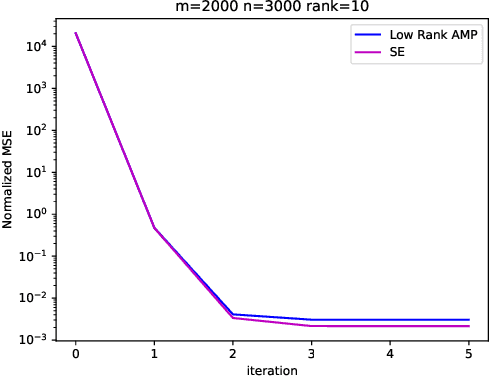
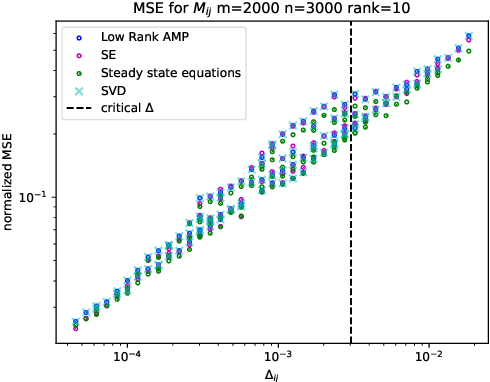
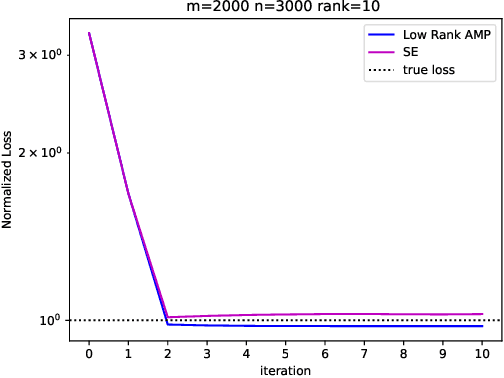
Abstract:Embeddings are a basic initial feature extraction step in many machine learning models, particularly in natural language processing. An embedding attempts to map data tokens to a low-dimensional space where similar tokens are mapped to vectors that are close to one another by some metric in the embedding space. A basic question is how well can such embedding be learned? To study this problem, we consider a simple probability model for discrete data where there is some "true" but unknown embedding where the correlation of random variables is related to the similarity of the embeddings. Under this model, it is shown that the embeddings can be learned by a variant of low-rank approximate message passing (AMP) method. The AMP approach enables precise predictions of the accuracy of the estimation in certain high-dimensional limits. In particular, the methodology provides insight on the relations of key parameters such as the number of samples per value, the frequency of the terms, and the strength of the embedding correlation on the probability distribution. Our theoretical findings are validated by simulations on both synthetic data and real text data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge