Joel Stremmel
Give me Some Hard Questions: Synthetic Data Generation for Clinical QA
Dec 05, 2024



Abstract:Clinical Question Answering (QA) systems enable doctors to quickly access patient information from electronic health records (EHRs). However, training these systems requires significant annotated data, which is limited due to the expertise needed and the privacy concerns associated with clinical data. This paper explores generating Clinical QA data using large language models (LLMs) in a zero-shot setting. We find that naive prompting often results in easy questions that do not reflect the complexity of clinical scenarios. To address this, we propose two prompting strategies: 1) instructing the model to generate questions that do not overlap with the input context, and 2) summarizing the input record using a predefined schema to scaffold question generation. Experiments on two Clinical QA datasets demonstrate that our method generates more challenging questions, significantly improving fine-tuning performance over baselines. We compare synthetic and gold data and find a gap between their training efficacy resulting from the quality of synthetically generated answers.
LLMs in Biomedicine: A study on clinical Named Entity Recognition
Apr 10, 2024



Abstract:Large Language Models (LLMs) demonstrate remarkable versatility in various NLP tasks but encounter distinct challenges in biomedicine due to medical language complexities and data scarcity. This paper investigates the application of LLMs in the medical domain by exploring strategies to enhance their performance for the Named-Entity Recognition (NER) task. Specifically, our study reveals the importance of meticulously designed prompts in biomedicine. Strategic selection of in-context examples yields a notable improvement, showcasing ~15-20\% increase in F1 score across all benchmark datasets for few-shot clinical NER. Additionally, our findings suggest that integrating external resources through prompting strategies can bridge the gap between general-purpose LLM proficiency and the specialized demands of medical NER. Leveraging a medical knowledge base, our proposed method inspired by Retrieval-Augmented Generation (RAG) can boost the F1 score of LLMs for zero-shot clinical NER. We will release the code upon publication.
XAIQA: Explainer-Based Data Augmentation for Extractive Question Answering
Dec 06, 2023Abstract:Extractive question answering (QA) systems can enable physicians and researchers to query medical records, a foundational capability for designing clinical studies and understanding patient medical history. However, building these systems typically requires expert-annotated QA pairs. Large language models (LLMs), which can perform extractive QA, depend on high quality data in their prompts, specialized for the application domain. We introduce a novel approach, XAIQA, for generating synthetic QA pairs at scale from data naturally available in electronic health records. Our method uses the idea of a classification model explainer to generate questions and answers about medical concepts corresponding to medical codes. In an expert evaluation with two physicians, our method identifies $2.2\times$ more semantic matches and $3.8\times$ more clinical abbreviations than two popular approaches that use sentence transformers to create QA pairs. In an ML evaluation, adding our QA pairs improves performance of GPT-4 as an extractive QA model, including on difficult questions. In both the expert and ML evaluations, we examine trade-offs between our method and sentence transformers for QA pair generation depending on question difficulty.
Surpassing GPT-4 Medical Coding with a Two-Stage Approach
Nov 22, 2023Abstract:Recent advances in large language models (LLMs) show potential for clinical applications, such as clinical decision support and trial recommendations. However, the GPT-4 LLM predicts an excessive number of ICD codes for medical coding tasks, leading to high recall but low precision. To tackle this challenge, we introduce LLM-codex, a two-stage approach to predict ICD codes that first generates evidence proposals using an LLM and then employs an LSTM-based verification stage. The LSTM learns from both the LLM's high recall and human expert's high precision, using a custom loss function. Our model is the only approach that simultaneously achieves state-of-the-art results in medical coding accuracy, accuracy on rare codes, and sentence-level evidence identification to support coding decisions without training on human-annotated evidence according to experiments on the MIMIC dataset.
Extend and Explain: Interpreting Very Long Language Models
Sep 07, 2022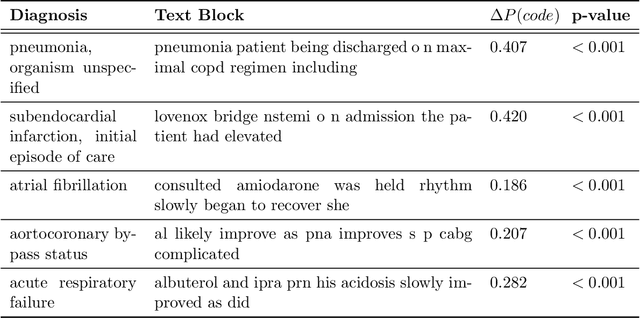
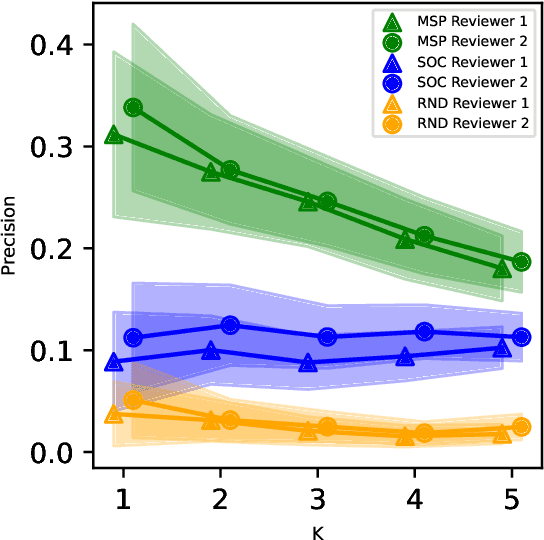
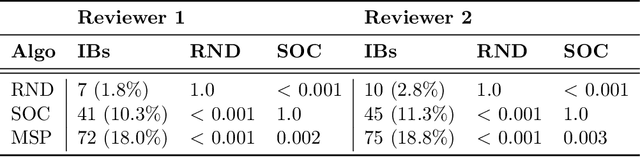
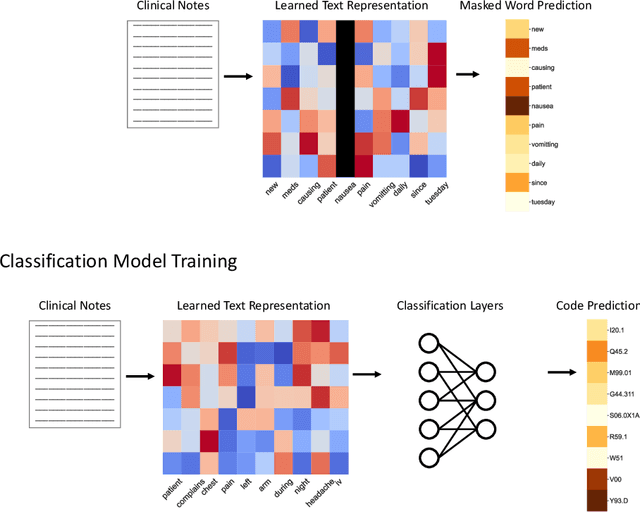
Abstract:While Transformer language models (LMs) are state-of-the-art for information extraction, long text introduces computational challenges requiring suboptimal preprocessing steps or alternative model architectures. Sparse-attention LMs can represent longer sequences, overcoming performance hurdles. However, it remains unclear how to explain predictions from these models, as not all tokens attend to each other in the self-attention layers, and long sequences pose computational challenges for explainability algorithms when runtime depends on document length. These challenges are severe in the medical context where documents can be very long, and machine learning (ML) models must be auditable and trustworthy. We introduce a novel Masked Sampling Procedure (MSP) to identify the text blocks that contribute to a prediction, apply MSP in the context of predicting diagnoses from medical text, and validate our approach with a blind review by two clinicians. Our method identifies about 1.7x more clinically informative text blocks than the previous state-of-the-art, runs up to 100x faster, and is tractable for generating important phrase pairs. MSP is particularly well-suited to long LMs but can be applied to any text classifier. We provide a general implementation of MSP.
Pretraining Federated Text Models for Next Word Prediction
May 12, 2020
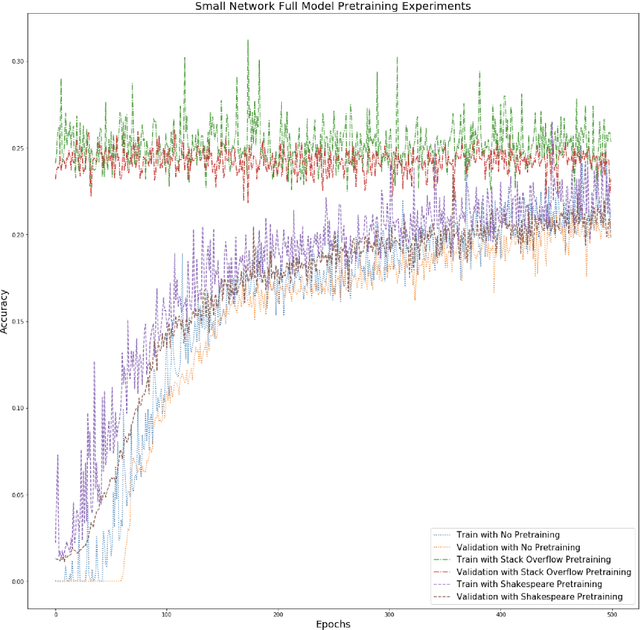
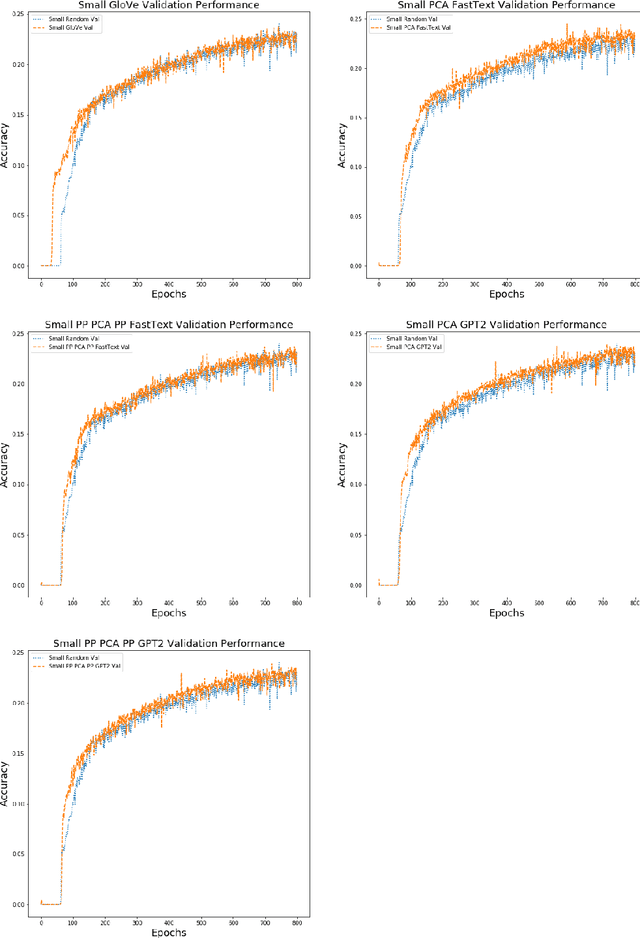
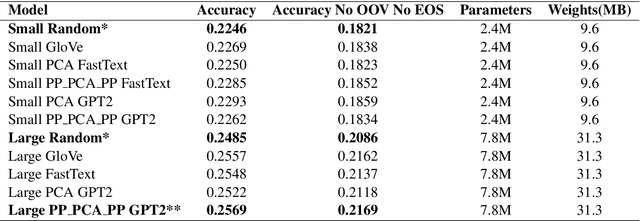
Abstract:Federated learning is a decentralized approach for training models on distributed devices, by summarizing local changes and sending aggregate parameters from local models to the cloud rather than the data itself. In this research we employ the idea of transfer learning to federated training for next word prediction (NWP) and conduct a number of experiments demonstrating enhancements to current baselines for which federated NWP models have been successful. Specifically, we compare federated training baselines from randomly initialized models to various combinations of pretraining approaches including pretrained word embeddings and whole model pretraining followed by federated fine tuning for NWP on a dataset of Stack Overflow posts. We realize lift in performance using pretrained embeddings without exacerbating the number of required training rounds or memory footprint. We also observe notable differences using centrally pretrained networks, especially depending on the datasets used. Our research offers effective, yet inexpensive, improvements to federated NWP and paves the way for more rigorous experimentation of transfer learning techniques for federated learning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge