Fan Bai
FIRE-Bench: Evaluating Agents on the Rediscovery of Scientific Insights
Feb 02, 2026Abstract:Autonomous agents powered by large language models (LLMs) promise to accelerate scientific discovery end-to-end, but rigorously evaluating their capacity for verifiable discovery remains a central challenge. Existing benchmarks face a trade-off: they either heavily rely on LLM-as-judge evaluations of automatically generated research outputs or optimize convenient yet isolated performance metrics that provide coarse proxies for scientific insight. To address this gap, we introduce FIRE-Bench (Full-cycle Insight Rediscovery Evaluation), a benchmark that evaluates agents through the rediscovery of established findings from recent, high-impact machine learning research. Agents are given only a high-level research question extracted from a published, verified study and must autonomously explore ideas, design experiments, implement code, execute their plans, and derive conclusions supported by empirical evidence. We evaluate a range of state-of-the-art agents with frontier LLMs backbones like gpt-5 on FIRE-Bench. Our results show that full-cycle scientific research remains challenging for current agent systems: even the strongest agents achieve limited rediscovery success (<50 F1), exhibit high variance across runs, and display recurring failure modes in experimental design, execution, and evidence-based reasoning. FIRE-Bench provides a rigorous and diagnostic framework for measuring progress toward reliable agent-driven scientific discovery.
What Is Seen Cannot Be Unseen: The Disruptive Effect of Knowledge Conflict on Large Language Models
Jun 06, 2025Abstract:Large language models frequently rely on both contextual input and parametric knowledge to perform tasks. However, these sources can come into conflict, especially when retrieved documents contradict the model's parametric knowledge. We propose a diagnostic framework to systematically evaluate LLM behavior under context-memory conflict, where the contextual information diverges from their parametric beliefs. We construct diagnostic data that elicit these conflicts and analyze model performance across multiple task types. Our findings reveal that (1) knowledge conflict has minimal impact on tasks that do not require knowledge utilization, (2) model performance is consistently higher when contextual and parametric knowledge are aligned, (3) models are unable to fully suppress their internal knowledge even when instructed, and (4) providing rationales that explain the conflict increases reliance on contexts. These insights raise concerns about the validity of model-based evaluation and underscore the need to account for knowledge conflict in the deployment of LLMs.
Label-Guided In-Context Learning for Named Entity Recognition
May 29, 2025Abstract:In-context learning (ICL) enables large language models (LLMs) to perform new tasks using only a few demonstrations. In Named Entity Recognition (NER), demonstrations are typically selected based on semantic similarity to the test instance, ignoring training labels and resulting in suboptimal performance. We introduce DEER, a new method that leverages training labels through token-level statistics to improve ICL performance. DEER first enhances example selection with a label-guided, token-based retriever that prioritizes tokens most informative for entity recognition. It then prompts the LLM to revisit error-prone tokens, which are also identified using label statistics, and make targeted corrections. Evaluated on five NER datasets using four different LLMs, DEER consistently outperforms existing ICL methods and approaches the performance of supervised fine-tuning. Further analysis shows its effectiveness on both seen and unseen entities and its robustness in low-resource settings.
Give me Some Hard Questions: Synthetic Data Generation for Clinical QA
Dec 05, 2024



Abstract:Clinical Question Answering (QA) systems enable doctors to quickly access patient information from electronic health records (EHRs). However, training these systems requires significant annotated data, which is limited due to the expertise needed and the privacy concerns associated with clinical data. This paper explores generating Clinical QA data using large language models (LLMs) in a zero-shot setting. We find that naive prompting often results in easy questions that do not reflect the complexity of clinical scenarios. To address this, we propose two prompting strategies: 1) instructing the model to generate questions that do not overlap with the input context, and 2) summarizing the input record using a predefined schema to scaffold question generation. Experiments on two Clinical QA datasets demonstrate that our method generates more challenging questions, significantly improving fine-tuning performance over baselines. We compare synthetic and gold data and find a gap between their training efficacy resulting from the quality of synthetically generated answers.
Touchstone Benchmark: Are We on the Right Way for Evaluating AI Algorithms for Medical Segmentation?
Nov 06, 2024



Abstract:How can we test AI performance? This question seems trivial, but it isn't. Standard benchmarks often have problems such as in-distribution and small-size test sets, oversimplified metrics, unfair comparisons, and short-term outcome pressure. As a consequence, good performance on standard benchmarks does not guarantee success in real-world scenarios. To address these problems, we present Touchstone, a large-scale collaborative segmentation benchmark of 9 types of abdominal organs. This benchmark is based on 5,195 training CT scans from 76 hospitals around the world and 5,903 testing CT scans from 11 additional hospitals. This diverse test set enhances the statistical significance of benchmark results and rigorously evaluates AI algorithms across various out-of-distribution scenarios. We invited 14 inventors of 19 AI algorithms to train their algorithms, while our team, as a third party, independently evaluated these algorithms on three test sets. In addition, we also evaluated pre-existing AI frameworks--which, differing from algorithms, are more flexible and can support different algorithms--including MONAI from NVIDIA, nnU-Net from DKFZ, and numerous other open-source frameworks. We are committed to expanding this benchmark to encourage more innovation of AI algorithms for the medical domain.
M3D: Advancing 3D Medical Image Analysis with Multi-Modal Large Language Models
Mar 31, 2024



Abstract:Medical image analysis is essential to clinical diagnosis and treatment, which is increasingly supported by multi-modal large language models (MLLMs). However, previous research has primarily focused on 2D medical images, leaving 3D images under-explored, despite their richer spatial information. This paper aims to advance 3D medical image analysis with MLLMs. To this end, we present a large-scale 3D multi-modal medical dataset, M3D-Data, comprising 120K image-text pairs and 662K instruction-response pairs specifically tailored for various 3D medical tasks, such as image-text retrieval, report generation, visual question answering, positioning, and segmentation. Additionally, we propose M3D-LaMed, a versatile multi-modal large language model for 3D medical image analysis. Furthermore, we introduce a new 3D multi-modal medical benchmark, M3D-Bench, which facilitates automatic evaluation across eight tasks. Through comprehensive evaluation, our method proves to be a robust model for 3D medical image analysis, outperforming existing solutions. All code, data, and models are publicly available at: https://github.com/BAAI-DCAI/M3D.
SAMv2: A Unified Framework for Learning Appearance, Semantic and Cross-Modality Anatomical Embeddings
Nov 28, 2023



Abstract:Identifying anatomical structures (e.g., lesions or landmarks) in medical images plays a fundamental role in medical image analysis. As an exemplar-based landmark detection method, Self-supervised Anatomical eMbedding (SAM) learns a discriminative embedding for each voxel in the image and has shown promising results on various tasks. However, SAM still faces challenges in: (1) differentiating voxels with similar appearance but different semantic meanings (\textit{e.g.}, two adjacent structures without clear borders); (2) matching voxels with similar semantics but markedly different appearance (e.g., the same vessel before and after contrast injection); and (3) cross-modality matching (e.g., CT-MRI registration). To overcome these challenges, we propose SAMv2, which is a unified framework designed to learn appearance, semantic, and cross-modality anatomical embeddings. Specifically, SAMv2 incorporates three key innovations: (1) semantic embedding learning with prototypical contrastive loss; (2) a fixed-point-based matching strategy; and (3) an iterative approach for cross-modality embedding learning. We thoroughly evaluated SAMv2 across three tasks, including one-shot landmark detection, lesion tracking on longitudinal CT scans, and CT-MRI affine/rigid registration with varying field of view. Our results suggest that SAMv2 outperforms SAM and other state-of-the-art methods, offering a robust and versatile approach for landmark based medical image analysis tasks. Code and trained models are available at: https://github.com/alibaba-damo-academy/self-supervised-anatomical-embedding-v2
SegVol: Universal and Interactive Volumetric Medical Image Segmentation
Nov 22, 2023



Abstract:Precise image segmentation provides clinical study with meaningful and well-structured information. Despite the remarkable progress achieved in medical image segmentation, there is still an absence of foundation segmentation model that can segment a wide range of anatomical categories with easy user interaction. In this paper, we propose a universal and interactive volumetric medical image segmentation model, named SegVol. By training on 90k unlabeled Computed Tomography (CT) volumes and 6k labeled CTs, this foundation model supports the segmentation of over 200 anatomical categories using semantic and spatial prompts. Extensive experiments verify that SegVol outperforms the state of the art by a large margin on multiple segmentation benchmarks. Notably, on three challenging lesion datasets, our method achieves around 20% higher Dice score than nnU-Net. The model and data are publicly available at: https://github.com/BAAI-DCAI/SegVol.
PromptAgent: Strategic Planning with Language Models Enables Expert-level Prompt Optimization
Oct 25, 2023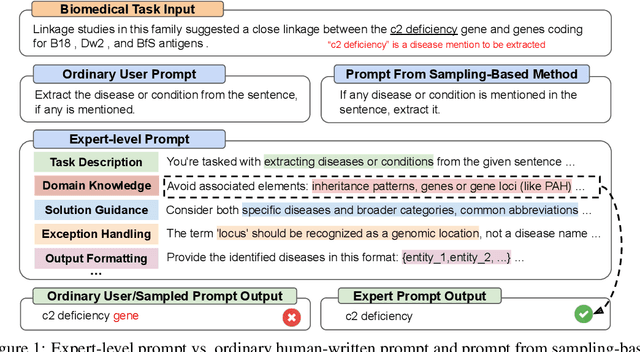
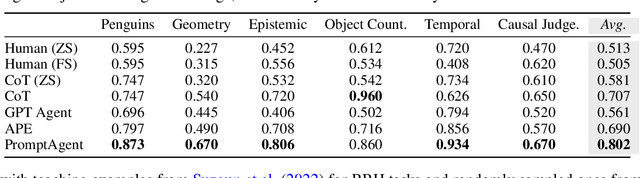
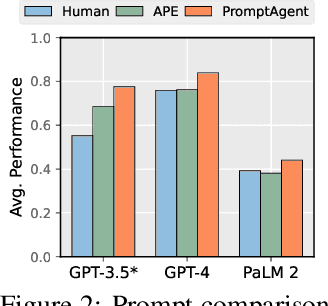
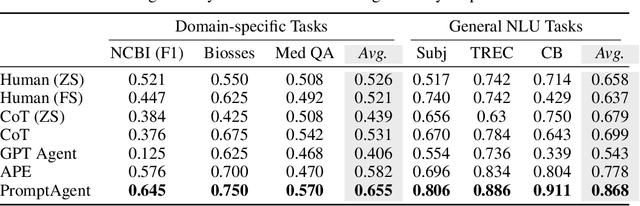
Abstract:Highly effective, task-specific prompts are often heavily engineered by experts to integrate detailed instructions and domain insights based on a deep understanding of both instincts of large language models (LLMs) and the intricacies of the target task. However, automating the generation of such expert-level prompts remains elusive. Existing prompt optimization methods tend to overlook the depth of domain knowledge and struggle to efficiently explore the vast space of expert-level prompts. Addressing this, we present PromptAgent, an optimization method that autonomously crafts prompts equivalent in quality to those handcrafted by experts. At its core, PromptAgent views prompt optimization as a strategic planning problem and employs a principled planning algorithm, rooted in Monte Carlo tree search, to strategically navigate the expert-level prompt space. Inspired by human-like trial-and-error exploration, PromptAgent induces precise expert-level insights and in-depth instructions by reflecting on model errors and generating constructive error feedback. Such a novel framework allows the agent to iteratively examine intermediate prompts (states), refine them based on error feedbacks (actions), simulate future rewards, and search for high-reward paths leading to expert prompts. We apply PromptAgent to 12 tasks spanning three practical domains: BIG-Bench Hard (BBH), as well as domain-specific and general NLP tasks, showing it significantly outperforms strong Chain-of-Thought and recent prompt optimization baselines. Extensive analyses emphasize its capability to craft expert-level, detailed, and domain-insightful prompts with great efficiency and generalizability.
A Consumer-tier based Visual-Brain Machine Interface for Augmented Reality Glasses Interactions
Aug 29, 2023



Abstract:Objective.Visual-Brain Machine Interface(V-BMI) has provide a novel interaction technique for Augmented Reality (AR) industries. Several state-of-arts work has demonstates its high accuracy and real-time interaction capbilities. However, most of the studies employ EEGs devices that are rigid and difficult to apply in real-life AR glasseses application sceniraros. Here we develop a consumer-tier Visual-Brain Machine Inteface(V-BMI) system specialized for Augmented Reality(AR) glasses interactions. Approach. The developed system consists of a wearable hardware which takes advantages of fast set-up, reliable recording and comfortable wearable experience that specificized for AR glasses applications. Complementing this hardware, we have devised a software framework that facilitates real-time interactions within the system while accommodating a modular configuration to enhance scalability. Main results. The developed hardware is only 110g and 120x85x23 mm, which with 1 Tohm and peak to peak voltage is less than 1.5 uV, and a V-BMI based angry bird game and an Internet of Thing (IoT) AR applications are deisgned, we demonstrated such technology merits of intuitive experience and efficiency interaction. The real-time interaction accuracy is between 85 and 96 percentages in a commercial AR glasses (DTI is 2.24s and ITR 65 bits-min ). Significance. Our study indicates the developed system can provide an essential hardware-software framework for consumer based V-BMI AR glasses. Also, we derive several pivotal design factors for a consumer-grade V-BMI-based AR system: 1) Dynamic adaptation of stimulation patterns-classification methods via computer vision algorithms is necessary for AR glasses applications; and 2) Algorithmic localization to foster system stability and latency reduction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge