Eric P. Xing
CellMaster: Collaborative Cell Type Annotation in Single-Cell Analysis
Feb 12, 2026Abstract:Single-cell RNA-seq (scRNA-seq) enables atlas-scale profiling of complex tissues, revealing rare lineages and transient states. Yet, assigning biologically valid cell identities remains a bottleneck because markers are tissue- and state-dependent, and novel states lack references. We present CellMaster, an AI agent that mimics expert practice for zero-shot cell-type annotation. Unlike existing automated tools, CellMaster leverages LLM-encoded knowledge (e.g., GPT-4o) to perform on-the-fly annotation with interpretable rationales, without pre-training or fixed marker databases. Across 9 datasets spanning 8 tissues, CellMaster improved accuracy by 7.1% over best-performing baselines (including CellTypist and scTab) in automatic mode. With human-in-the-loop refinement, this advantage increased to 18.6%, with a 22.1% gain on subtype populations. The system demonstrates particular strength in rare and novel cell states where baselines often fail. Source code and the web application are available at \href{https://github.com/AnonymousGym/CellMaster}{https://github.com/AnonymousGym/CellMaster}.
scPilot: Large Language Model Reasoning Toward Automated Single-Cell Analysis and Discovery
Feb 12, 2026Abstract:We present scPilot, the first systematic framework to practice omics-native reasoning: a large language model (LLM) converses in natural language while directly inspecting single-cell RNA-seq data and on-demand bioinformatics tools. scPilot converts core single-cell analyses, i.e., cell-type annotation, developmental-trajectory reconstruction, and transcription-factor targeting, into step-by-step reasoning problems that the model must solve, justify, and, when needed, revise with new evidence. To measure progress, we release scBench, a suite of 9 expertly curated datasets and graders that faithfully evaluate the omics-native reasoning capability of scPilot w.r.t various LLMs. Experiments with o1 show that iterative omics-native reasoning lifts average accuracy by 11% for cell-type annotation and Gemini-2.5-Pro cuts trajectory graph-edit distance by 30% versus one-shot prompting, while generating transparent reasoning traces explain marker gene ambiguity and regulatory logic. By grounding LLMs in raw omics data, scPilot enables auditable, interpretable, and diagnostically informative single-cell analyses. Code, data, and package are available at https://github.com/maitrix-org/scPilot
Retrieval-Aware Distillation for Transformer-SSM Hybrids
Feb 11, 2026Abstract:State-space models (SSMs) offer efficient sequence modeling but lag behind Transformers on benchmarks that require in-context retrieval. Prior work links this gap to a small set of attention heads, termed Gather-and-Aggregate (G&A), which SSMs struggle to reproduce. We propose *retrieval-aware distillation*, which converts a pretrained Transformer into a hybrid student by preserving only these retrieval-critical heads and distilling the rest into recurrent heads. We identify the essential heads via ablation on a synthetic retrieval task, producing a hybrid with sparse, non-uniform attention placement. We show that preserving **just 2% of attention heads recovers over 95% of teacher performance on retrieval-heavy tasks** (10 heads in a 1B model), requiring far fewer heads than hybrids that retain at least 25%. We further find that large recurrent states often compensate for missing retrieval: once retrieval is handled by these heads, the SSM backbone can be simplified with limited loss, even with an $8\times$ reduction in state dimension. By reducing both the attention cache and the SSM state, the resulting hybrid is $5$--$6\times$ more memory-efficient than comparable hybrids, closing the Transformer--SSM gap at a fraction of the memory cost.
FIRE-Bench: Evaluating Agents on the Rediscovery of Scientific Insights
Feb 02, 2026Abstract:Autonomous agents powered by large language models (LLMs) promise to accelerate scientific discovery end-to-end, but rigorously evaluating their capacity for verifiable discovery remains a central challenge. Existing benchmarks face a trade-off: they either heavily rely on LLM-as-judge evaluations of automatically generated research outputs or optimize convenient yet isolated performance metrics that provide coarse proxies for scientific insight. To address this gap, we introduce FIRE-Bench (Full-cycle Insight Rediscovery Evaluation), a benchmark that evaluates agents through the rediscovery of established findings from recent, high-impact machine learning research. Agents are given only a high-level research question extracted from a published, verified study and must autonomously explore ideas, design experiments, implement code, execute their plans, and derive conclusions supported by empirical evidence. We evaluate a range of state-of-the-art agents with frontier LLMs backbones like gpt-5 on FIRE-Bench. Our results show that full-cycle scientific research remains challenging for current agent systems: even the strongest agents achieve limited rediscovery success (<50 F1), exhibit high variance across runs, and display recurring failure modes in experimental design, execution, and evidence-based reasoning. FIRE-Bench provides a rigorous and diagnostic framework for measuring progress toward reliable agent-driven scientific discovery.
"Rebuilding" Statistics in the Age of AI: A Town Hall Discussion on Culture, Infrastructure, and Training
Jan 24, 2026Abstract:This article presents the full, original record of the 2024 Joint Statistical Meetings (JSM) town hall, "Statistics in the Age of AI," which convened leading statisticians to discuss how the field is evolving in response to advances in artificial intelligence, foundation models, large-scale empirical modeling, and data-intensive infrastructures. The town hall was structured around open panel discussion and extensive audience Q&A, with the aim of eliciting candid, experience-driven perspectives rather than formal presentations or prepared statements. This document preserves the extended exchanges among panelists and audience members, with minimal editorial intervention, and organizes the conversation around five recurring questions concerning disciplinary culture and practices, data curation and "data work," engagement with modern empirical modeling, training for large-scale AI applications, and partnerships with key AI stakeholders. By providing an archival record of this discussion, the preprint aims to support transparency, community reflection, and ongoing dialogue about the evolving role of statistics in the data- and AI-centric future.
Bridging Streaming Continual Learning via In-Context Large Tabular Models
Dec 12, 2025Abstract:In streaming scenarios, models must learn continuously, adapting to concept drifts without erasing previously acquired knowledge. However, existing research communities address these challenges in isolation. Continual Learning (CL) focuses on long-term retention and mitigating catastrophic forgetting, often without strict real-time constraints. Stream Learning (SL) emphasizes rapid, efficient adaptation to high-frequency data streams, but typically neglects forgetting. Recent efforts have tried to combine these paradigms, yet no clear algorithmic overlap exists. We argue that large in-context tabular models (LTMs) provide a natural bridge for Streaming Continual Learning (SCL). In our view, unbounded streams should be summarized on-the-fly into compact sketches that can be consumed by LTMs. This recovers the classical SL motivation of compressing massive streams with fixed-size guarantees, while simultaneously aligning with the experience-replay desiderata of CL. To clarify this bridge, we show how the SL and CL communities implicitly adopt a divide-to-conquer strategy to manage the tension between plasticity (performing well on the current distribution) and stability (retaining past knowledge), while also imposing a minimal complexity constraint that motivates diversification (avoiding redundancy in what is stored) and retrieval (re-prioritizing past information when needed). Within this perspective, we propose structuring SCL with LTMs around two core principles of data selection for in-context learning: (1) distribution matching, which balances plasticity and stability, and (2) distribution compression, which controls memory size through diversification and retrieval mechanisms.
Beyond the Black Box: Identifiable Interpretation and Control in Generative Models via Causal Minimality
Dec 11, 2025Abstract:Deep generative models, while revolutionizing fields like image and text generation, largely operate as opaque black boxes, hindering human understanding, control, and alignment. While methods like sparse autoencoders (SAEs) show remarkable empirical success, they often lack theoretical guarantees, risking subjective insights. Our primary objective is to establish a principled foundation for interpretable generative models. We demonstrate that the principle of causal minimality -- favoring the simplest causal explanation -- can endow the latent representations of diffusion vision and autoregressive language models with clear causal interpretation and robust, component-wise identifiable control. We introduce a novel theoretical framework for hierarchical selection models, where higher-level concepts emerge from the constrained composition of lower-level variables, better capturing the complex dependencies in data generation. Under theoretically derived minimality conditions (manifesting as sparsity or compression constraints), we show that learned representations can be equivalent to the true latent variables of the data-generating process. Empirically, applying these constraints to leading generative models allows us to extract their innate hierarchical concept graphs, offering fresh insights into their internal knowledge organization. Furthermore, these causally grounded concepts serve as levers for fine-grained model steering, paving the way for transparent, reliable systems.
PAN: A World Model for General, Interactable, and Long-Horizon World Simulation
Nov 15, 2025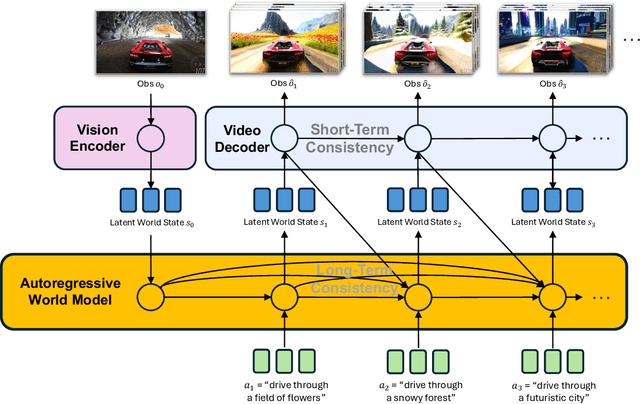
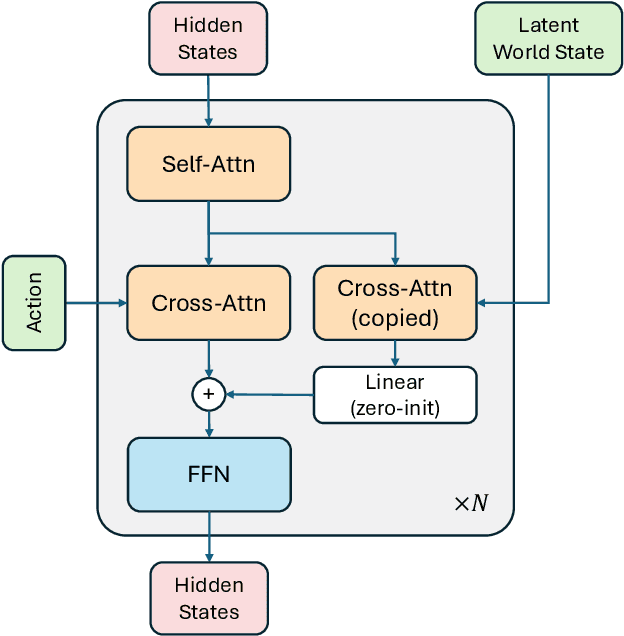
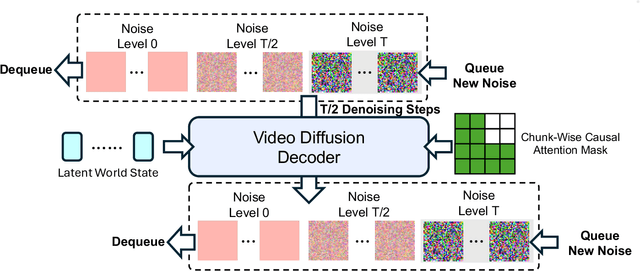
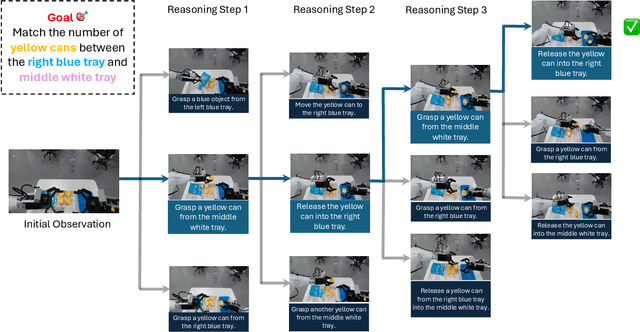
Abstract:A world model enables an intelligent agent to imagine, predict, and reason about how the world evolves in response to its actions, and accordingly to plan and strategize. While recent video generation models produce realistic visual sequences, they typically operate in the prompt-to-full-video manner without causal control, interactivity, or long-horizon consistency required for purposeful reasoning. Existing world modeling efforts, on the other hand, often focus on restricted domains (e.g., physical, game, or 3D-scene dynamics) with limited depth and controllability, and struggle to generalize across diverse environments and interaction formats. In this work, we introduce PAN, a general, interactable, and long-horizon world model that predicts future world states through high-quality video simulation conditioned on history and natural language actions. PAN employs the Generative Latent Prediction (GLP) architecture that combines an autoregressive latent dynamics backbone based on a large language model (LLM), which grounds simulation in extensive text-based knowledge and enables conditioning on language-specified actions, with a video diffusion decoder that reconstructs perceptually detailed and temporally coherent visual observations, to achieve a unification between latent space reasoning (imagination) and realizable world dynamics (reality). Trained on large-scale video-action pairs spanning diverse domains, PAN supports open-domain, action-conditioned simulation with coherent, long-term dynamics. Extensive experiments show that PAN achieves strong performance in action-conditioned world simulation, long-horizon forecasting, and simulative reasoning compared to other video generators and world models, taking a step towards general world models that enable predictive simulation of future world states for reasoning and acting.
Step-Aware Policy Optimization for Reasoning in Diffusion Large Language Models
Oct 02, 2025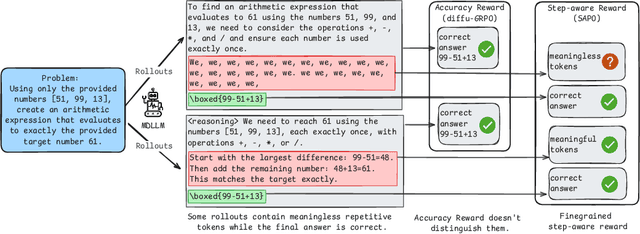
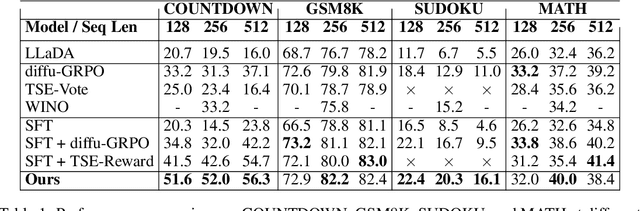
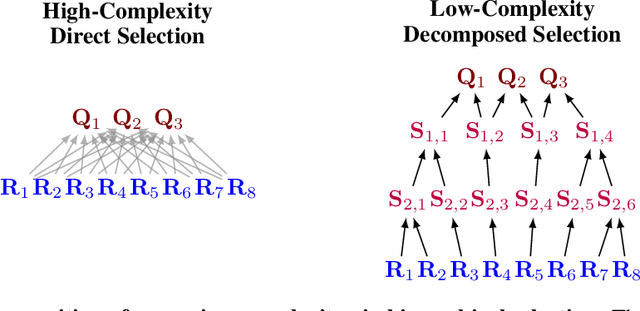
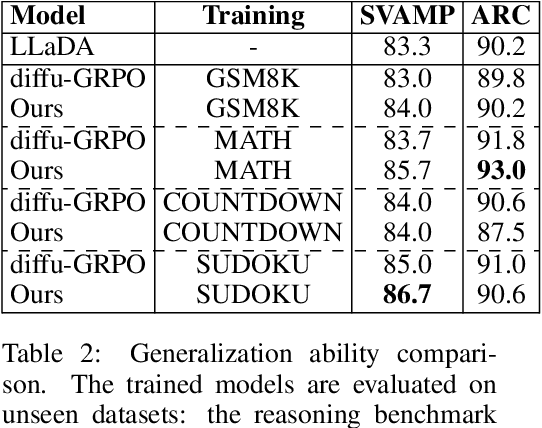
Abstract:Diffusion language models (dLLMs) offer a promising, non-autoregressive paradigm for text generation, yet training them for complex reasoning remains a key challenge. Current reinforcement learning approaches often rely on sparse, outcome-based rewards, which can reinforce flawed reasoning paths that lead to coincidentally correct answers. We argue that this stems from a fundamental mismatch with the natural structure of reasoning. We first propose a theoretical framework that formalizes complex problem solving as a hierarchical selection process, where an intractable global constraint is decomposed into a series of simpler, localized logical steps. This framework provides a principled foundation for algorithm design, including theoretical insights into the identifiability of this latent reasoning structure. Motivated by this theory, we identify unstructured refinement -- a failure mode where a model's iterative steps do not contribute meaningfully to the solution -- as a core deficiency in existing methods. We then introduce Step-Aware Policy Optimization (SAPO), a novel RL algorithm that aligns the dLLM's denoising process with the latent reasoning hierarchy. By using a process-based reward function that encourages incremental progress, SAPO guides the model to learn structured, coherent reasoning paths. Our empirical results show that this principled approach significantly improves performance on challenging reasoning benchmarks and enhances the interpretability of the generation process.
K2-Think: A Parameter-Efficient Reasoning System
Sep 09, 2025Abstract:K2-Think is a reasoning system that achieves state-of-the-art performance with a 32B parameter model, matching or surpassing much larger models like GPT-OSS 120B and DeepSeek v3.1. Built on the Qwen2.5 base model, our system shows that smaller models can compete at the highest levels by combining advanced post-training and test-time computation techniques. The approach is based on six key technical pillars: Long Chain-of-thought Supervised Finetuning, Reinforcement Learning with Verifiable Rewards (RLVR), Agentic planning prior to reasoning, Test-time Scaling, Speculative Decoding, and Inference-optimized Hardware, all using publicly available open-source datasets. K2-Think excels in mathematical reasoning, achieving state-of-the-art scores on public benchmarks for open-source models, while also performing strongly in other areas such as Code and Science. Our results confirm that a more parameter-efficient model like K2-Think 32B can compete with state-of-the-art systems through an integrated post-training recipe that includes long chain-of-thought training and strategic inference-time enhancements, making open-source reasoning systems more accessible and affordable. K2-Think is freely available at k2think.ai, offering best-in-class inference speeds of over 2,000 tokens per second per request via the Cerebras Wafer-Scale Engine.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge