Xiaoli Yin
Non-Contrast CT Esophageal Varices Grading through Clinical Prior-Enhanced Multi-Organ Analysis
Dec 22, 2025Abstract:Esophageal varices (EV) represent a critical complication of portal hypertension, affecting approximately 60% of cirrhosis patients with a significant bleeding risk of ~30%. While traditionally diagnosed through invasive endoscopy, non-contrast computed tomography (NCCT) presents a potential non-invasive alternative that has yet to be fully utilized in clinical practice. We present Multi-Organ-COhesion Network++ (MOON++), a novel multimodal framework that enhances EV assessment through comprehensive analysis of NCCT scans. Inspired by clinical evidence correlating organ volumetric relationships with liver disease severity, MOON++ synthesizes imaging characteristics of the esophagus, liver, and spleen through multimodal learning. We evaluated our approach using 1,631 patients, those with endoscopically confirmed EV were classified into four severity grades. Validation in 239 patient cases and independent testing in 289 cases demonstrate superior performance compared to conventional single organ methods, achieving an AUC of 0.894 versus 0.803 for the severe grade EV classification (G3 versus <G3) and 0.921 versus 0.793 for the differentiation of moderate to severe grades (>=G2 versus <G2). We conducted a reader study involving experienced radiologists to further validate the performance of MOON++. To our knowledge, MOON++ represents the first comprehensive multi-organ NCCT analysis framework incorporating clinical knowledge priors for EV assessment, potentially offering a promising non-invasive diagnostic alternative.
Improved Esophageal Varices Assessment from Non-Contrast CT Scans
Jul 18, 2024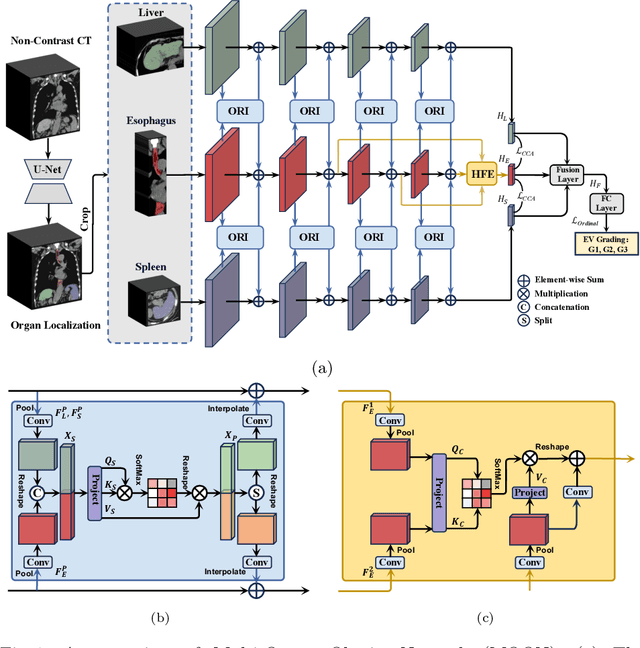
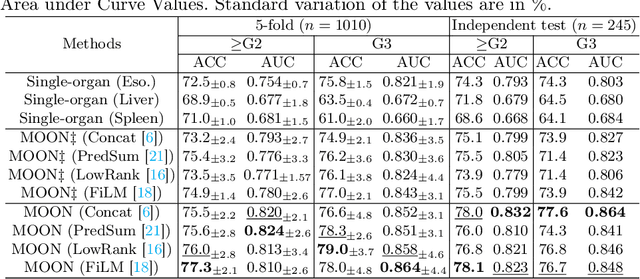
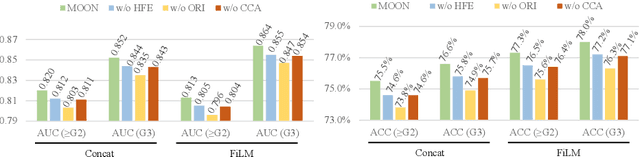
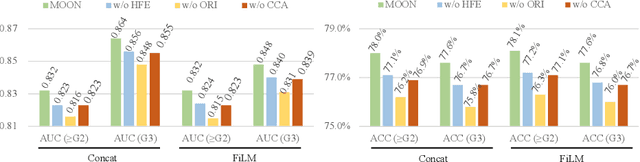
Abstract:Esophageal varices (EV), a serious health concern resulting from portal hypertension, are traditionally diagnosed through invasive endoscopic procedures. Despite non-contrast computed tomography (NC-CT) imaging being a less expensive and non-invasive imaging modality, it has yet to gain full acceptance as a primary clinical diagnostic tool for EV evaluation. To overcome existing diagnostic challenges, we present the Multi-Organ-cOhesion-Network (MOON), a novel framework enhancing the analysis of critical organ features in NC-CT scans for effective assessment of EV. Drawing inspiration from the thorough assessment practices of radiologists, MOON establishes a cohesive multiorgan analysis model that unifies the imaging features of the related organs of EV, namely esophagus, liver, and spleen. This integration significantly increases the diagnostic accuracy for EV. We have compiled an extensive NC-CT dataset of 1,255 patients diagnosed with EV, spanning three grades of severity. Each case is corroborated by endoscopic diagnostic results. The efficacy of MOON has been substantiated through a validation process involving multi-fold cross-validation on 1,010 cases and an independent test on 245 cases, exhibiting superior diagnostic performance compared to methods focusing solely on the esophagus (for classifying severe grade: AUC of 0.864 versus 0.803, and for moderate to severe grades: AUC of 0.832 versus 0.793). To our knowledge, MOON is the first work to incorporate a synchronized multi-organ NC-CT analysis for EV assessment, providing a more acceptable and minimally invasive alternative for patients compared to traditional endoscopy.
LIDIA: Precise Liver Tumor Diagnosis on Multi-Phase Contrast-Enhanced CT via Iterative Fusion and Asymmetric Contrastive Learning
Jul 18, 2024



Abstract:The early detection and precise diagnosis of liver tumors are tasks of critical clinical value, yet they pose significant challenges due to the high heterogeneity and variability of liver tumors. In this work, a precise LIver tumor DIAgnosis network on multi-phase contrast-enhance CT, named LIDIA, is proposed for real-world scenario. To fully utilize all available phases in contrast-enhanced CT, LIDIA first employs the iterative fusion module to aggregate variable numbers of image phases, thereby capturing the features of lesions at different phases for better tumor diagnosis. To effectively mitigate the high heterogeneity problem of liver tumors, LIDIA incorporates asymmetric contrastive learning to enhance the discriminability between different classes. To evaluate our method, we constructed a large-scale dataset comprising 1,921 patients and 8,138 lesions. LIDIA has achieved an average AUC of 93.6% across eight different types of lesions, demonstrating its effectiveness. Besides, LIDIA also demonstrated strong generalizability with an average AUC of 89.3% when tested on an external cohort of 828 patients.
Modality-Agnostic Structural Image Representation Learning for Deformable Multi-Modality Medical Image Registration
Feb 29, 2024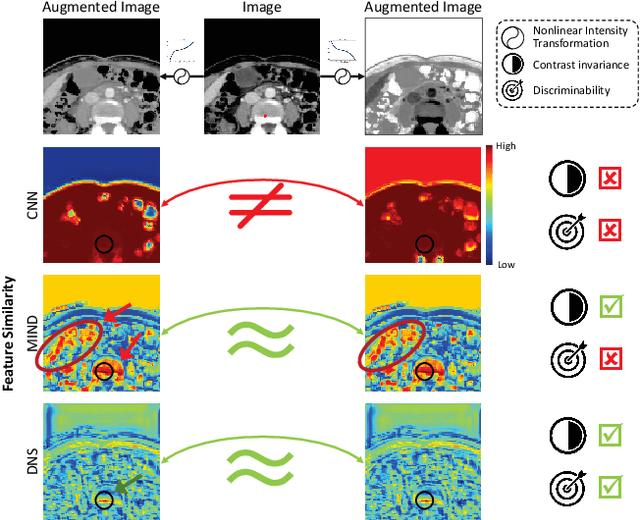
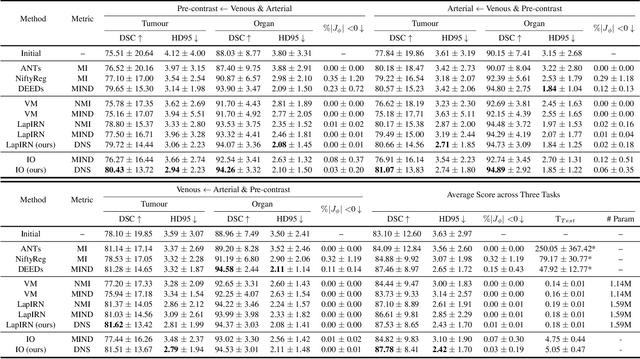
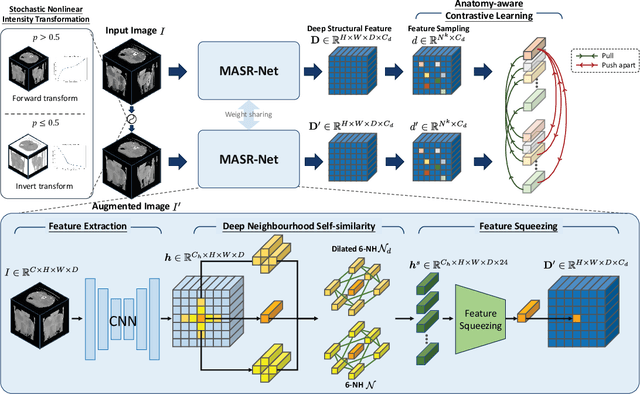
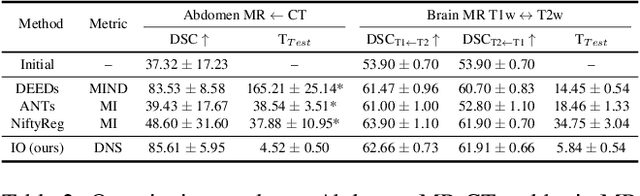
Abstract:Establishing dense anatomical correspondence across distinct imaging modalities is a foundational yet challenging procedure for numerous medical image analysis studies and image-guided radiotherapy. Existing multi-modality image registration algorithms rely on statistical-based similarity measures or local structural image representations. However, the former is sensitive to locally varying noise, while the latter is not discriminative enough to cope with complex anatomical structures in multimodal scans, causing ambiguity in determining the anatomical correspondence across scans with different modalities. In this paper, we propose a modality-agnostic structural representation learning method, which leverages Deep Neighbourhood Self-similarity (DNS) and anatomy-aware contrastive learning to learn discriminative and contrast-invariance deep structural image representations (DSIR) without the need for anatomical delineations or pre-aligned training images. We evaluate our method on multiphase CT, abdomen MR-CT, and brain MR T1w-T2w registration. Comprehensive results demonstrate that our method is superior to the conventional local structural representation and statistical-based similarity measures in terms of discriminability and accuracy.
SLPT: Selective Labeling Meets Prompt Tuning on Label-Limited Lesion Segmentation
Aug 09, 2023


Abstract:Medical image analysis using deep learning is often challenged by limited labeled data and high annotation costs. Fine-tuning the entire network in label-limited scenarios can lead to overfitting and suboptimal performance. Recently, prompt tuning has emerged as a more promising technique that introduces a few additional tunable parameters as prompts to a task-agnostic pre-trained model, and updates only these parameters using supervision from limited labeled data while keeping the pre-trained model unchanged. However, previous work has overlooked the importance of selective labeling in downstream tasks, which aims to select the most valuable downstream samples for annotation to achieve the best performance with minimum annotation cost. To address this, we propose a framework that combines selective labeling with prompt tuning (SLPT) to boost performance in limited labels. Specifically, we introduce a feature-aware prompt updater to guide prompt tuning and a TandEm Selective LAbeling (TESLA) strategy. TESLA includes unsupervised diversity selection and supervised selection using prompt-based uncertainty. In addition, we propose a diversified visual prompt tuning strategy to provide multi-prompt-based discrepant predictions for TESLA. We evaluate our method on liver tumor segmentation and achieve state-of-the-art performance, outperforming traditional fine-tuning with only 6% of tunable parameters, also achieving 94% of full-data performance by labeling only 5% of the data.
Liver Tumor Screening and Diagnosis in CT with Pixel-Lesion-Patient Network
Jul 17, 2023Abstract:Liver tumor segmentation and classification are important tasks in computer aided diagnosis. We aim to address three problems: liver tumor screening and preliminary diagnosis in non-contrast computed tomography (CT), and differential diagnosis in dynamic contrast-enhanced CT. A novel framework named Pixel-Lesion-pAtient Network (PLAN) is proposed. It uses a mask transformer to jointly segment and classify each lesion with improved anchor queries and a foreground-enhanced sampling loss. It also has an image-wise classifier to effectively aggregate global information and predict patient-level diagnosis. A large-scale multi-phase dataset is collected containing 939 tumor patients and 810 normal subjects. 4010 tumor instances of eight types are extensively annotated. On the non-contrast tumor screening task, PLAN achieves 95% and 96% in patient-level sensitivity and specificity. On contrast-enhanced CT, our lesion-level detection precision, recall, and classification accuracy are 92%, 89%, and 86%, outperforming widely used CNN and transformers for lesion segmentation. We also conduct a reader study on a holdout set of 250 cases. PLAN is on par with a senior human radiologist, showing the clinical significance of our results.
Devil is in the Queries: Advancing Mask Transformers for Real-world Medical Image Segmentation and Out-of-Distribution Localization
Apr 01, 2023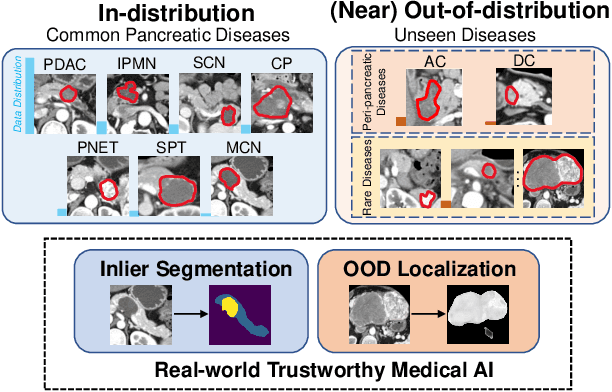
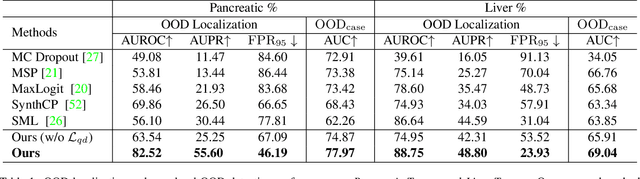
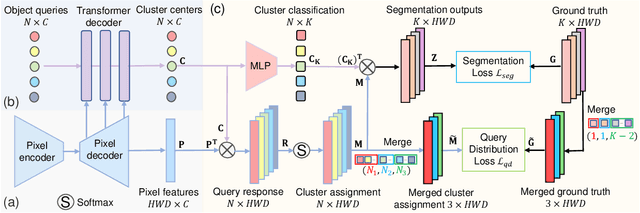
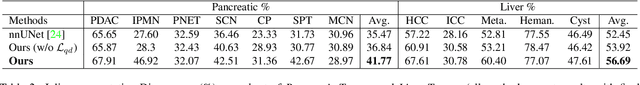
Abstract:Real-world medical image segmentation has tremendous long-tailed complexity of objects, among which tail conditions correlate with relatively rare diseases and are clinically significant. A trustworthy medical AI algorithm should demonstrate its effectiveness on tail conditions to avoid clinically dangerous damage in these out-of-distribution (OOD) cases. In this paper, we adopt the concept of object queries in Mask Transformers to formulate semantic segmentation as a soft cluster assignment. The queries fit the feature-level cluster centers of inliers during training. Therefore, when performing inference on a medical image in real-world scenarios, the similarity between pixels and the queries detects and localizes OOD regions. We term this OOD localization as MaxQuery. Furthermore, the foregrounds of real-world medical images, whether OOD objects or inliers, are lesions. The difference between them is less than that between the foreground and background, possibly misleading the object queries to focus redundantly on the background. Thus, we propose a query-distribution (QD) loss to enforce clear boundaries between segmentation targets and other regions at the query level, improving the inlier segmentation and OOD indication. Our proposed framework is tested on two real-world segmentation tasks, i.e., segmentation of pancreatic and liver tumors, outperforming previous state-of-the-art algorithms by an average of 7.39% on AUROC, 14.69% on AUPR, and 13.79% on FPR95 for OOD localization. On the other hand, our framework improves the performance of inlier segmentation by an average of 5.27% DSC when compared with the leading baseline nnUNet.
Towards a Single Unified Model for Effective Detection, Segmentation, and Diagnosis of Eight Major Cancers Using a Large Collection of CT Scans
Jan 28, 2023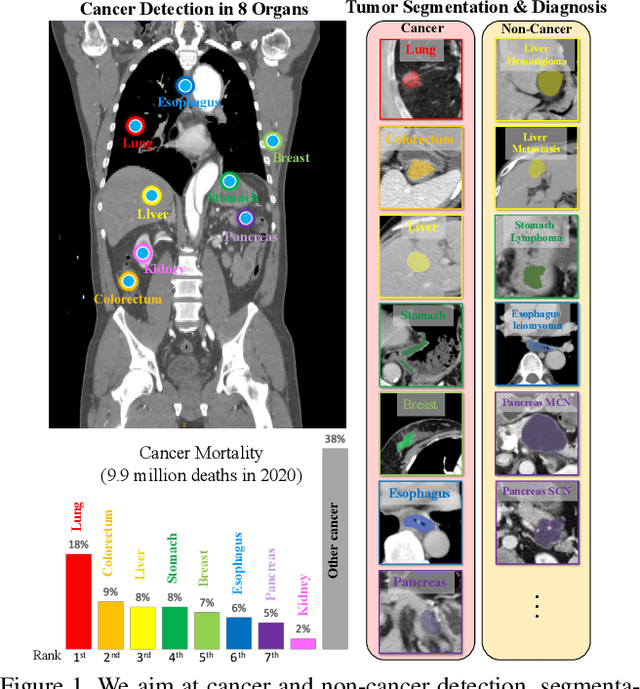
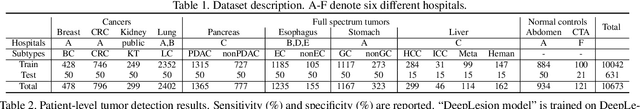
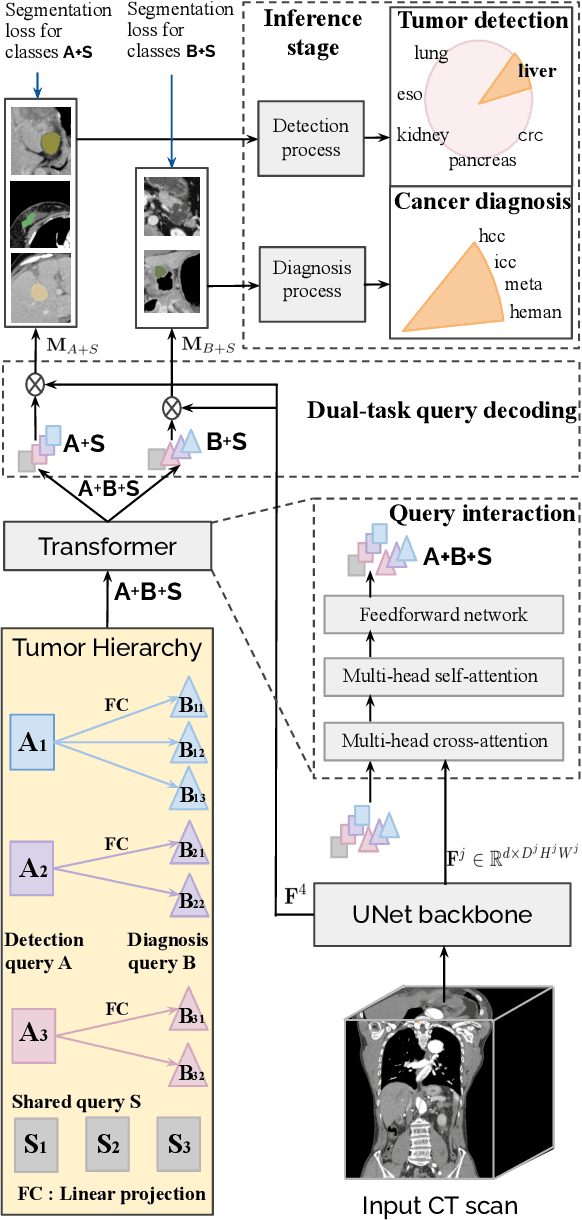
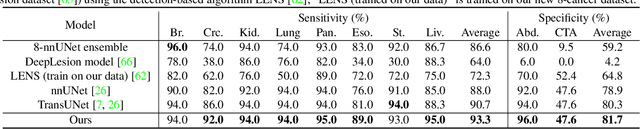
Abstract:Human readers or radiologists routinely perform full-body multi-organ multi-disease detection and diagnosis in clinical practice, while most medical AI systems are built to focus on single organs with a narrow list of a few diseases. This might severely limit AI's clinical adoption. A certain number of AI models need to be assembled non-trivially to match the diagnostic process of a human reading a CT scan. In this paper, we construct a Unified Tumor Transformer (UniT) model to detect (tumor existence and location) and diagnose (tumor characteristics) eight major cancer-prevalent organs in CT scans. UniT is a query-based Mask Transformer model with the output of multi-organ and multi-tumor semantic segmentation. We decouple the object queries into organ queries, detection queries and diagnosis queries, and further establish hierarchical relationships among the three groups. This clinically-inspired architecture effectively assists inter- and intra-organ representation learning of tumors and facilitates the resolution of these complex, anatomically related multi-organ cancer image reading tasks. UniT is trained end-to-end using a curated large-scale CT images of 10,042 patients including eight major types of cancers and occurring non-cancer tumors (all are pathology-confirmed with 3D tumor masks annotated by radiologists). On the test set of 631 patients, UniT has demonstrated strong performance under a set of clinically relevant evaluation metrics, substantially outperforming both multi-organ segmentation methods and an assembly of eight single-organ expert models in tumor detection, segmentation, and diagnosis. Such a unified multi-cancer image reading model (UniT) can significantly reduce the number of false positives produced by combined multi-system models. This moves one step closer towards a universal high-performance cancer screening tool.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge