Zifan Chen
AI-Driven Automated Tool for Abdominal CT Body Composition Analysis in Gastrointestinal Cancer Management
Mar 10, 2025Abstract:The incidence of gastrointestinal cancers remains significantly high, particularly in China, emphasizing the importance of accurate prognostic assessments and effective treatment strategies. Research shows a strong correlation between abdominal muscle and fat tissue composition and patient outcomes. However, existing manual methods for analyzing abdominal tissue composition are time-consuming and costly, limiting clinical research scalability. To address these challenges, we developed an AI-driven tool for automated analysis of abdominal CT scans to effectively identify and segment muscle, subcutaneous fat, and visceral fat. Our tool integrates a multi-view localization model and a high-precision 2D nnUNet-based segmentation model, demonstrating a localization accuracy of 90% and a Dice Score Coefficient of 0.967 for segmentation. Furthermore, it features an interactive interface that allows clinicians to refine the segmentation results, ensuring high-quality outcomes effectively. Our tool offers a standardized method for effectively extracting critical abdominal tissues, potentially enhancing the management and treatment for gastrointestinal cancers. The code is available at https://github.com/NanXinyu/AI-Tool4Abdominal-Seg.git}{https://github.com/NanXinyu/AI-Tool4Abdominal-Seg.git.
PropSAM: A Propagation-Based Model for Segmenting Any 3D Objects in Multi-Modal Medical Images
Aug 25, 2024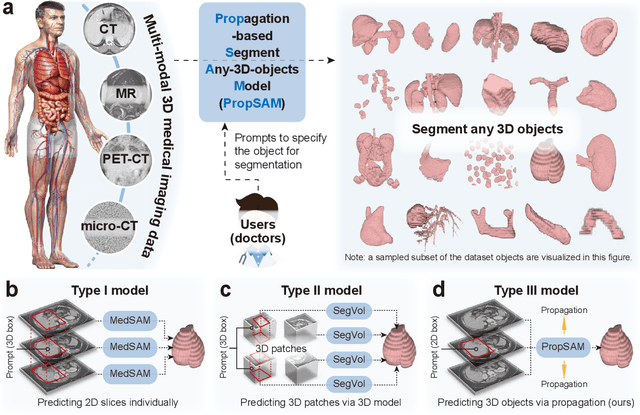
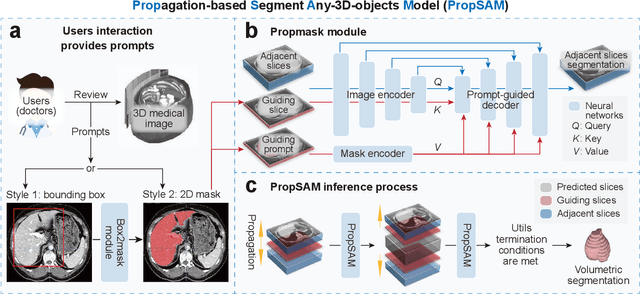
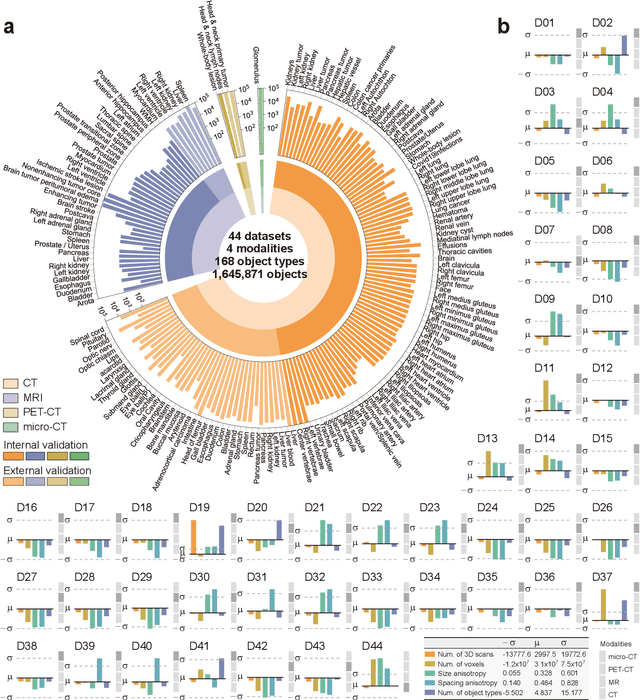
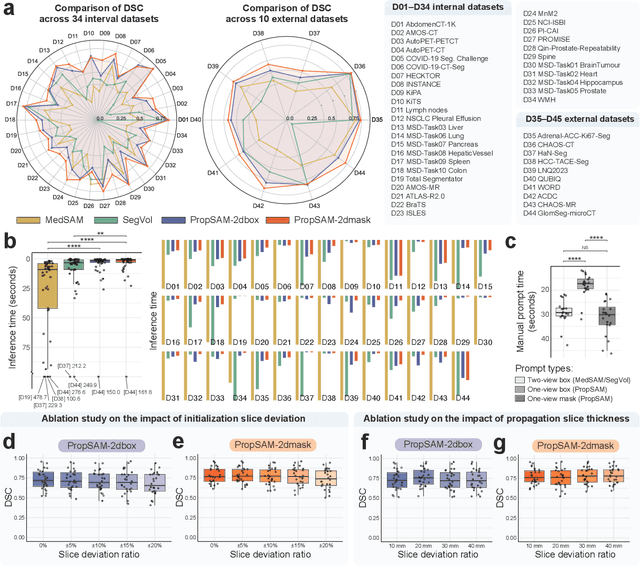
Abstract:Volumetric segmentation is crucial for medical imaging but is often constrained by labor-intensive manual annotations and the need for scenario-specific model training. Furthermore, existing general segmentation models are inefficient due to their design and inferential approaches. Addressing this clinical demand, we introduce PropSAM, a propagation-based segmentation model that optimizes the use of 3D medical structure information. PropSAM integrates a CNN-based UNet for intra-slice processing with a Transformer-based module for inter-slice propagation, focusing on structural and semantic continuities to enhance segmentation across various modalities. Distinctively, PropSAM operates on a one-view prompt, such as a 2D bounding box or sketch mask, unlike conventional models that require two-view prompts. It has demonstrated superior performance, significantly improving the Dice Similarity Coefficient (DSC) across 44 medical datasets and various imaging modalities, outperforming models like MedSAM and SegVol with an average DSC improvement of 18.1%. PropSAM also maintains stable predictions despite prompt deviations and varying propagation configurations, confirmed by one-way ANOVA tests with P>0.5985 and P>0.6131, respectively. Moreover, PropSAM's efficient architecture enables faster inference speeds (Wilcoxon rank-sum test, P<0.001) and reduces user interaction time by 37.8% compared to two-view prompt models. Its ability to handle irregular and complex objects with robust performance further demonstrates its potential in clinical settings, facilitating more automated and reliable medical imaging analyses with minimal retraining.
Large Language Models Illuminate a Progressive Pathway to Artificial Healthcare Assistant: A Review
Nov 03, 2023



Abstract:With the rapid development of artificial intelligence, large language models (LLMs) have shown promising capabilities in mimicking human-level language comprehension and reasoning. This has sparked significant interest in applying LLMs to enhance various aspects of healthcare, ranging from medical education to clinical decision support. However, medicine involves multifaceted data modalities and nuanced reasoning skills, presenting challenges for integrating LLMs. This paper provides a comprehensive review on the applications and implications of LLMs in medicine. It begins by examining the fundamental applications of general-purpose and specialized LLMs, demonstrating their utilities in knowledge retrieval, research support, clinical workflow automation, and diagnostic assistance. Recognizing the inherent multimodality of medicine, the review then focuses on multimodal LLMs, investigating their ability to process diverse data types like medical imaging and EHRs to augment diagnostic accuracy. To address LLMs' limitations regarding personalization and complex clinical reasoning, the paper explores the emerging development of LLM-powered autonomous agents for healthcare. Furthermore, it summarizes the evaluation methodologies for assessing LLMs' reliability and safety in medical contexts. Overall, this review offers an extensive analysis on the transformative potential of LLMs in modern medicine. It also highlights the pivotal need for continuous optimizations and ethical oversight before these models can be effectively integrated into clinical practice. Visit https://github.com/mingze-yuan/Awesome-LLM-Healthcare for an accompanying GitHub repository containing latest papers.
propnet: Propagating 2D Annotation to 3D Segmentation for Gastric Tumors on CT Scans
May 29, 2023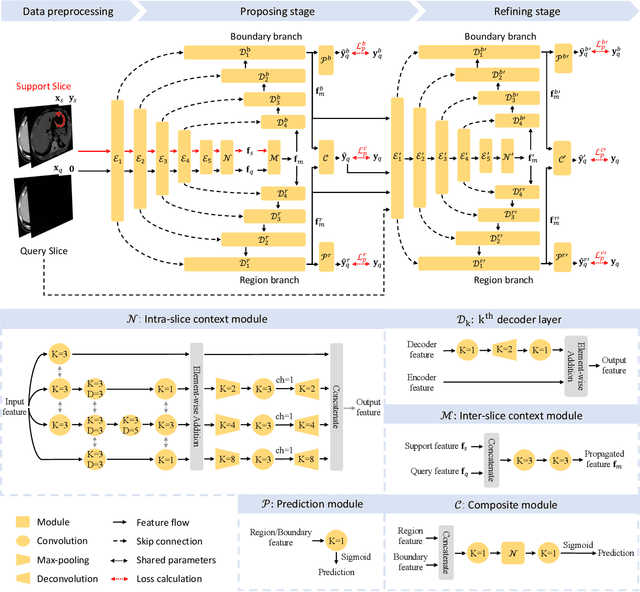
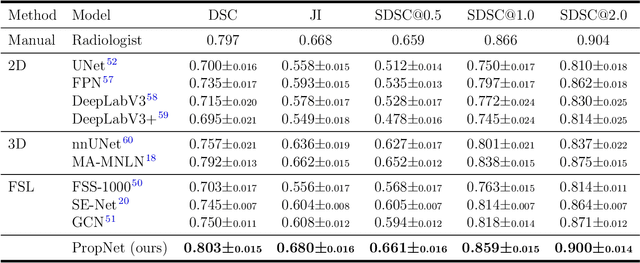
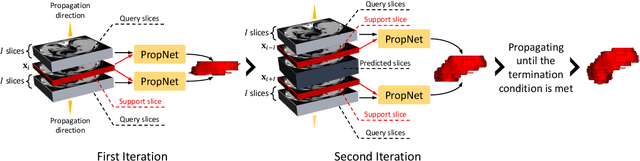
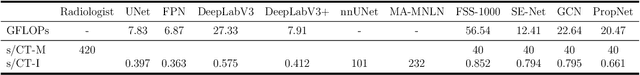
Abstract:**Background:** Accurate 3D CT scan segmentation of gastric tumors is pivotal for diagnosis and treatment. The challenges lie in the irregular shapes, blurred boundaries of tumors, and the inefficiency of existing methods. **Purpose:** We conducted a study to introduce a model, utilizing human-guided knowledge and unique modules, to address the challenges of 3D tumor segmentation. **Methods:** We developed the PropNet framework, propagating radiologists' knowledge from 2D annotations to the entire 3D space. This model consists of a proposing stage for coarse segmentation and a refining stage for improved segmentation, using two-way branches for enhanced performance and an up-down strategy for efficiency. **Results:** With 98 patient scans for training and 30 for validation, our method achieves a significant agreement with manual annotation (Dice of 0.803) and improves efficiency. The performance is comparable in different scenarios and with various radiologists' annotations (Dice between 0.785 and 0.803). Moreover, the model shows improved prognostic prediction performance (C-index of 0.620 vs. 0.576) on an independent validation set of 42 patients with advanced gastric cancer. **Conclusions:** Our model generates accurate tumor segmentation efficiently and stably, improving prognostic performance and reducing high-throughput image reading workload. This model can accelerate the quantitative analysis of gastric tumors and enhance downstream task performance.
Devil is in the Queries: Advancing Mask Transformers for Real-world Medical Image Segmentation and Out-of-Distribution Localization
Apr 01, 2023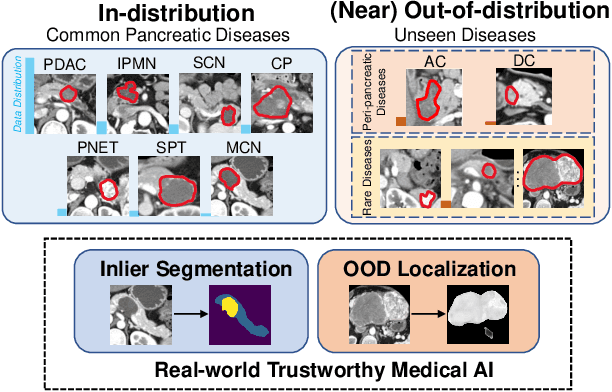
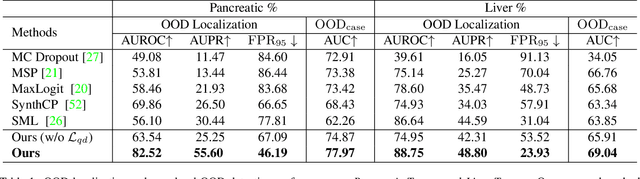
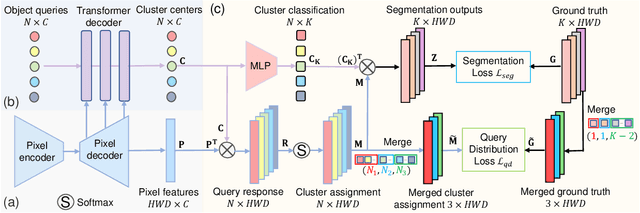
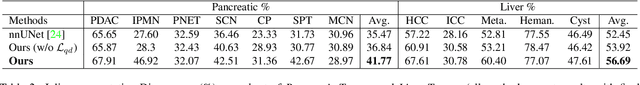
Abstract:Real-world medical image segmentation has tremendous long-tailed complexity of objects, among which tail conditions correlate with relatively rare diseases and are clinically significant. A trustworthy medical AI algorithm should demonstrate its effectiveness on tail conditions to avoid clinically dangerous damage in these out-of-distribution (OOD) cases. In this paper, we adopt the concept of object queries in Mask Transformers to formulate semantic segmentation as a soft cluster assignment. The queries fit the feature-level cluster centers of inliers during training. Therefore, when performing inference on a medical image in real-world scenarios, the similarity between pixels and the queries detects and localizes OOD regions. We term this OOD localization as MaxQuery. Furthermore, the foregrounds of real-world medical images, whether OOD objects or inliers, are lesions. The difference between them is less than that between the foreground and background, possibly misleading the object queries to focus redundantly on the background. Thus, we propose a query-distribution (QD) loss to enforce clear boundaries between segmentation targets and other regions at the query level, improving the inlier segmentation and OOD indication. Our proposed framework is tested on two real-world segmentation tasks, i.e., segmentation of pancreatic and liver tumors, outperforming previous state-of-the-art algorithms by an average of 7.39% on AUROC, 14.69% on AUPR, and 13.79% on FPR95 for OOD localization. On the other hand, our framework improves the performance of inlier segmentation by an average of 5.27% DSC when compared with the leading baseline nnUNet.
Region-Aware Metric Learning for Open World Semantic Segmentation via Meta-Channel Aggregation
May 17, 2022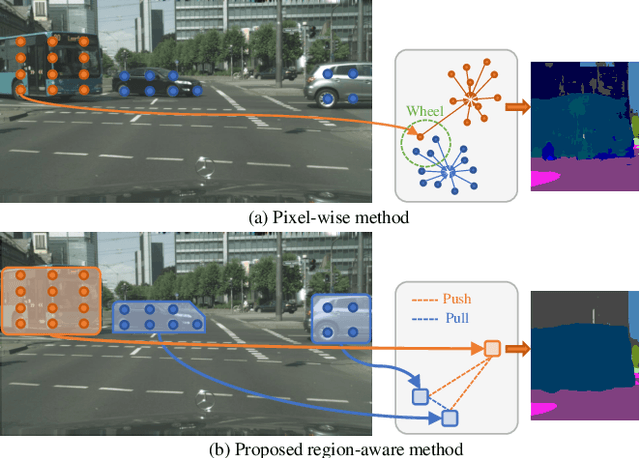
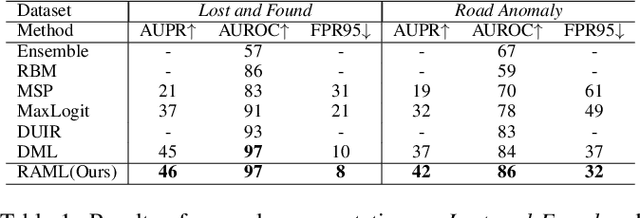
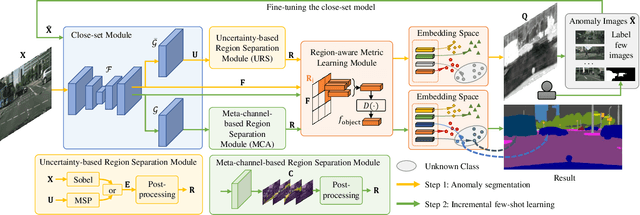
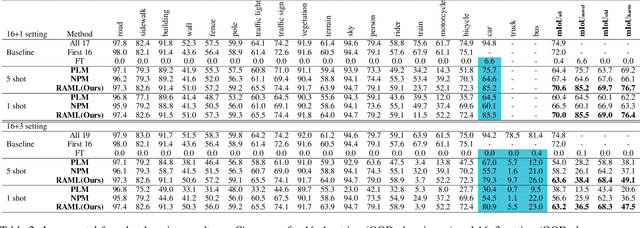
Abstract:As one of the most challenging and practical segmentation tasks, open-world semantic segmentation requires the model to segment the anomaly regions in the images and incrementally learn to segment out-of-distribution (OOD) objects, especially under a few-shot condition. The current state-of-the-art (SOTA) method, Deep Metric Learning Network (DMLNet), relies on pixel-level metric learning, with which the identification of similar regions having different semantics is difficult. Therefore, we propose a method called region-aware metric learning (RAML), which first separates the regions of the images and generates region-aware features for further metric learning. RAML improves the integrity of the segmented anomaly regions. Moreover, we propose a novel meta-channel aggregation (MCA) module to further separate anomaly regions, forming high-quality sub-region candidates and thereby improving the model performance for OOD objects. To evaluate the proposed RAML, we have conducted extensive experiments and ablation studies on Lost And Found and Road Anomaly datasets for anomaly segmentation and the CityScapes dataset for incremental few-shot learning. The results show that the proposed RAML achieves SOTA performance in both stages of open world segmentation. Our code and appendix are available at https://github.com/czifan/RAML.
Multi-Scale Context-Guided Lumbar Spine Disease Identification with Coarse-to-fine Localization and Classification
Mar 16, 2022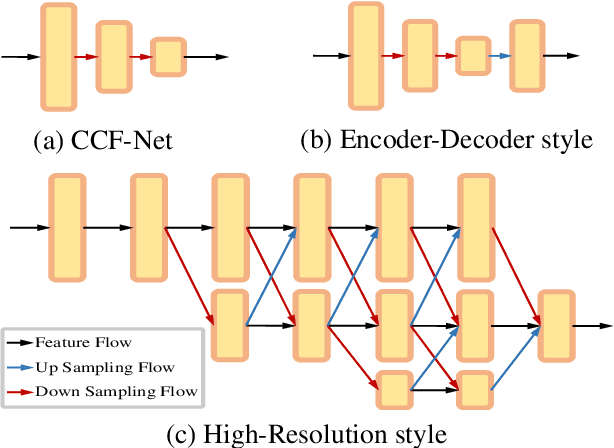
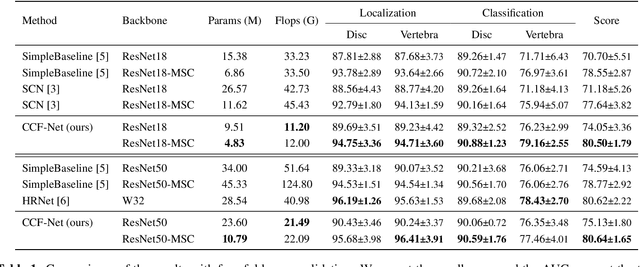
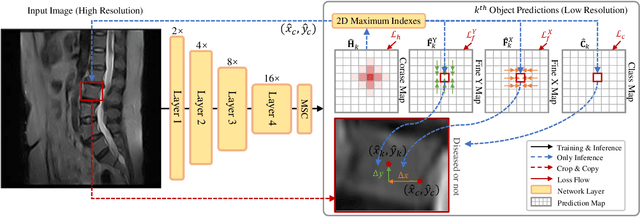
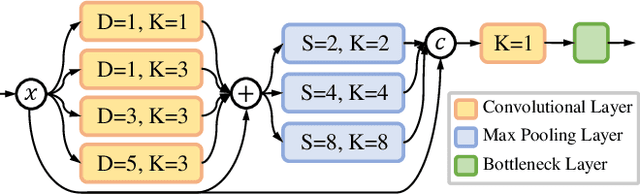
Abstract:Accurate and efficient lumbar spine disease identification is crucial for clinical diagnosis. However, existing deep learning models with millions of parameters often fail to learn with only hundreds or dozens of medical images. These models also ignore the contextual relationship between adjacent objects, such as between vertebras and intervertebral discs. This work introduces a multi-scale context-guided network with coarse-to-fine localization and classification, named CCF-Net, for lumbar spine disease identification. Specifically, in learning, we divide the localization objective into two parallel tasks, coarse and fine, which are more straightforward and effectively reduce the number of parameters and computational cost. The experimental results show that the coarse-to-fine design presents the potential to achieve high performance with fewer parameters and data requirements. Moreover, the multi-scale context-guided module can significantly improve the performance by 6.45% and 5.51% with ResNet18 and ResNet50, respectively. Our code is available at https://github.com/czifan/CCFNet.pytorch.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge