Hongfeng Li
Quantitative Analysis of Molecular Transport in the Extracellular Space Using Physics-Informed Neural Network
Jan 24, 2024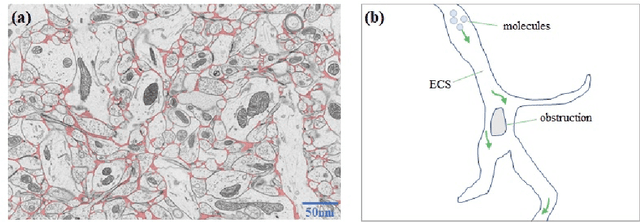



Abstract:The brain extracellular space (ECS), an irregular, extremely tortuous nanoscale space located between cells or between cells and blood vessels, is crucial for nerve cell survival. It plays a pivotal role in high-level brain functions such as memory, emotion, and sensation. However, the specific form of molecular transport within the ECS remain elusive. To address this challenge, this paper proposes a novel approach to quantitatively analyze the molecular transport within the ECS by solving an inverse problem derived from the advection-diffusion equation (ADE) using a physics-informed neural network (PINN). PINN provides a streamlined solution to the ADE without the need for intricate mathematical formulations or grid settings. Additionally, the optimization of PINN facilitates the automatic computation of the diffusion coefficient governing long-term molecule transport and the velocity of molecules driven by advection. Consequently, the proposed method allows for the quantitative analysis and identification of the specific pattern of molecular transport within the ECS through the calculation of the Peclet number. Experimental validation on two datasets of magnetic resonance images (MRIs) captured at different time points showcases the effectiveness of the proposed method. Notably, our simulations reveal identical molecular transport patterns between datasets representing rats with tracer injected into the same brain region. These findings highlight the potential of PINN as a promising tool for comprehensively exploring molecular transport within the ECS.
propnet: Propagating 2D Annotation to 3D Segmentation for Gastric Tumors on CT Scans
May 29, 2023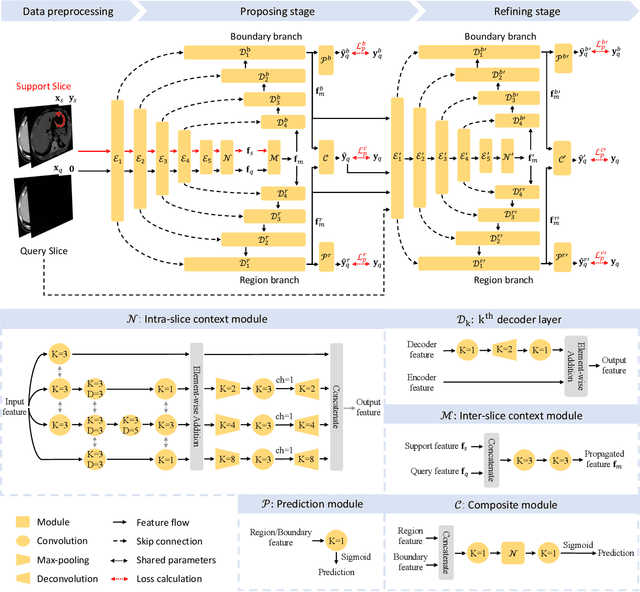
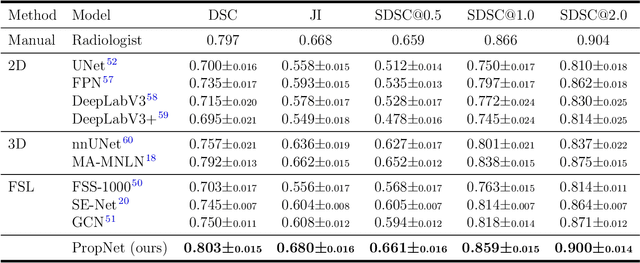
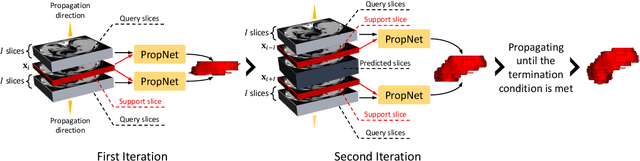
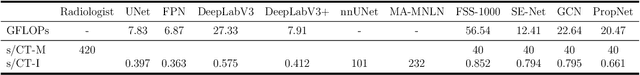
Abstract:**Background:** Accurate 3D CT scan segmentation of gastric tumors is pivotal for diagnosis and treatment. The challenges lie in the irregular shapes, blurred boundaries of tumors, and the inefficiency of existing methods. **Purpose:** We conducted a study to introduce a model, utilizing human-guided knowledge and unique modules, to address the challenges of 3D tumor segmentation. **Methods:** We developed the PropNet framework, propagating radiologists' knowledge from 2D annotations to the entire 3D space. This model consists of a proposing stage for coarse segmentation and a refining stage for improved segmentation, using two-way branches for enhanced performance and an up-down strategy for efficiency. **Results:** With 98 patient scans for training and 30 for validation, our method achieves a significant agreement with manual annotation (Dice of 0.803) and improves efficiency. The performance is comparable in different scenarios and with various radiologists' annotations (Dice between 0.785 and 0.803). Moreover, the model shows improved prognostic prediction performance (C-index of 0.620 vs. 0.576) on an independent validation set of 42 patients with advanced gastric cancer. **Conclusions:** Our model generates accurate tumor segmentation efficiently and stably, improving prognostic performance and reducing high-throughput image reading workload. This model can accelerate the quantitative analysis of gastric tumors and enhance downstream task performance.
Lightweight Dual-channel Target Speaker Separation for Mobile Voice Communication
Jun 05, 2021
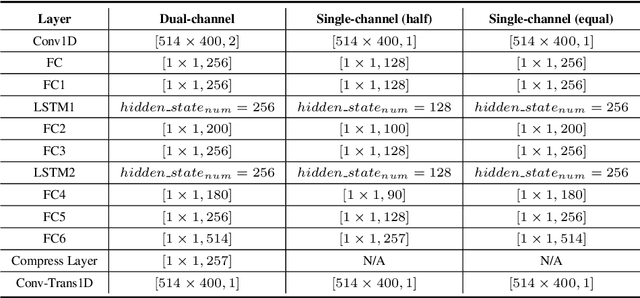
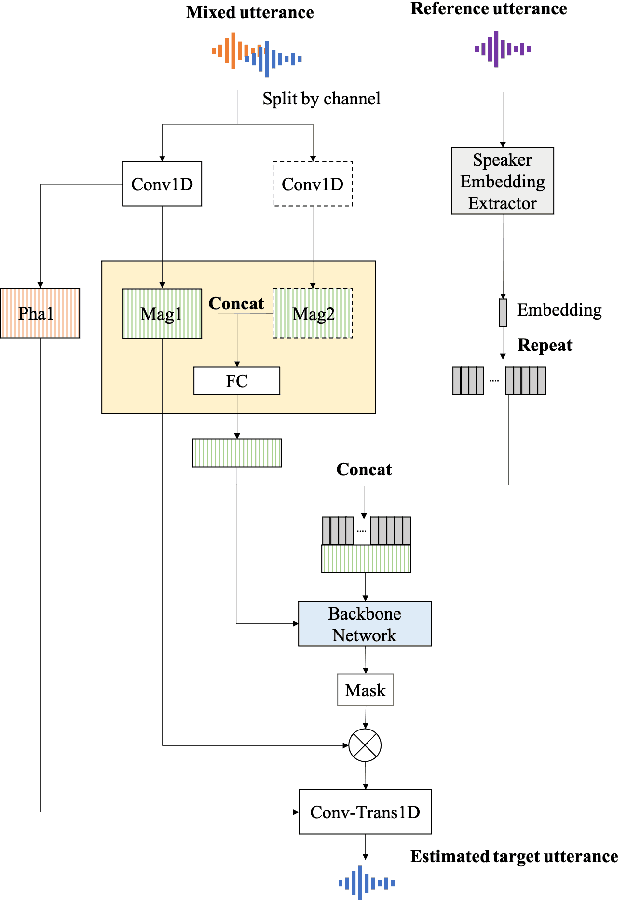
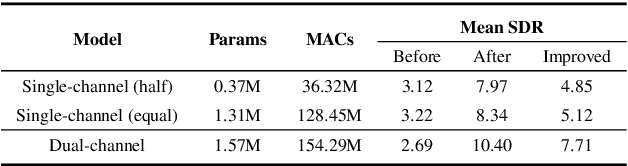
Abstract:Nowadays, there is a strong need to deploy the target speaker separation (TSS) model on mobile devices with a limitation of the model size and computational complexity. To better perform TSS for mobile voice communication, we first make a dual-channel dataset based on a specific scenario, LibriPhone. Specifically, to better mimic the real-case scenario, instead of simulating from the single-channel dataset, LibriPhone is made by simultaneously replaying pairs of utterances from LibriSpeech by two professional artificial heads and recording by two built-in microphones of the mobile. Then, we propose a lightweight time-frequency domain separation model, LSTM-Former, which is based on the LSTM framework with source-to-noise ratio (SI-SNR) loss. For the experiments on Libri-Phone, we explore the dual-channel LSTMFormer model and a single-channel version by a random single channel of Libri-Phone. Experimental result shows that the dual-channel LSTM-Former outperforms the single-channel LSTMFormer with relative 25% improvement. This work provides a feasible solution for the TSS task on mobile devices, playing back and recording multiple data sources in real application scenarios for getting dual-channel real data can assist the lightweight model to achieve higher performance.
Semi-supervised Sparse Representation with Graph Regularization for Image Classification
Nov 11, 2020



Abstract:Image classification is a challenging problem for computer in reality. Large numbers of methods can achieve satisfying performances with sufficient labeled images. However, labeled images are still highly limited for certain image classification tasks. Instead, lots of unlabeled images are available and easy to be obtained. Therefore, making full use of the available unlabeled data can be a potential way to further improve the performance of current image classification methods. In this paper, we propose a discriminative semi-supervised sparse representation algorithm for image classification. In the algorithm, the classification process is combined with the sparse coding to learn a data-driven linear classifier. To obtain discriminative predictions, the predicted labels are regularized with three graphs, i.e., the global manifold structure graph, the within-class graph and the between-classes graph. The constructed graphs are able to extract structure information included in both the labeled and unlabeled data. Moreover, the proposed method is extended to a kernel version for dealing with data that cannot be linearly classified. Accordingly, efficient algorithms are developed to solve the corresponding optimization problems. Experimental results on several challenging databases demonstrate that the proposed algorithm achieves excellent performances compared with related popular methods.
Skin disease diagnosis with deep learning: a review
Nov 11, 2020



Abstract:Skin cancer is one of the most threatening diseases worldwide. However, diagnosing a skin cancer correctly is challenging. Recently, deep learning algorithms have achieved excellent performance on various tasks. Particularly, they have been also implemented for the tasks of skin disease diagnosis. In this paper, we present a review on deep learning methods and their applications in skin disease diagnosis. We first introduce skin diseases and image acquisition methods in dermatology, and list several publicly available datasets for training and testing algorithms for skin disease diagnosis. Then, we introduce the conception of deep learning and review popular deep learning architectures. Thereafter, popular deep learning frameworks that facilitate the implementation of deep learning algorithms and performance evaluation metrics are presented. As an important part of this article, we then review the literatures involving deep learning methods for skin disease diagnosis from several aspects according to the specific tasks. Additionally, we discuss the challenges faced in the area of skin disease diagnosis with deep learning and suggest possible future research directions. Finally, we summarize the article. The major purpose of this article is to provide a conceptual and systematically review of the recent works on skin disease diagnosis with deep learning. Given the popularity of deep learning, there remains great challenges in the area, as well as opportunities that we can explore in the future.
PGU-net+: Progressive Growing of U-net+ for Automated Cervical Nuclei Segmentation
Nov 12, 2019

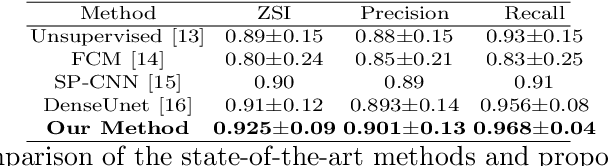

Abstract:Automated cervical nucleus segmentation based on deep learning can effectively improve the quantitative analysis of cervical cancer. However, accurate nuclei segmentation is still challenging. The classic U-net has not achieved satisfactory results on this task, because it mixes the information of different scales that affect each other, which limits the segmentation accuracy of the model. To solve this problem, we propose a progressive growing U-net (PGU-net+) model, which uses two paradigms to extract image features at different scales in a more independent way. First, we add residual modules between different scales of U-net, which enforces the model to learn the approximate shape of the annotation in the coarser scale, and to learn the residual between the annotation and the approximate shape in the finer scale. Second, we start to train the model with the coarsest part and then progressively add finer part to the training until the full model is included. When we train a finer part, we will reduce the learning rate of the previous coarser part, which further ensures that the model independently extracts information from different scales. We conduct several comparative experiments on the Herlev dataset. The experimental results show that the PGU-net+ has superior accuracy than the previous state-of-the-art methods on cervical nuclei segmentation.
Automated Segmentation of Cervical Nuclei in Pap Smear Images using Deformable Multi-path Ensemble Model
Dec 03, 2018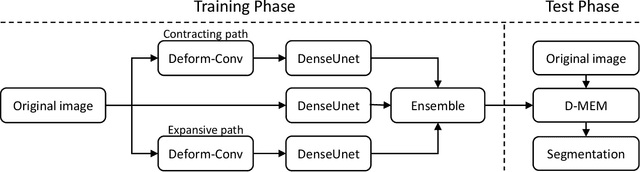
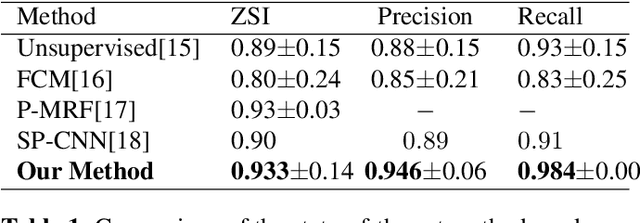
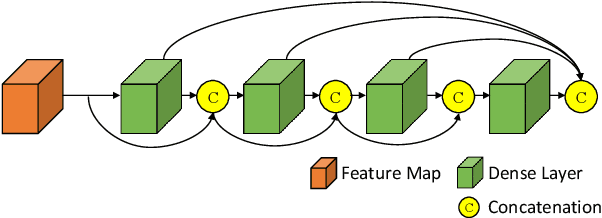
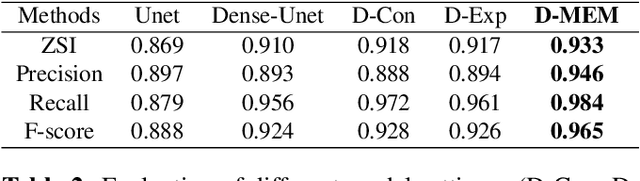
Abstract:Pap smear testing has been widely used for detecting cervical cancers based on the morphology properties of cell nuclei in microscopic image. An accurate nuclei segmentation could thus improve the success rate of cervical cancer screening. In this work, a method of automated cervical nuclei segmentation using Deformable Multipath Ensemble Model (D-MEM) is proposed. The approach adopts a U-shaped convolutional network as a backbone network, in which dense blocks are used to transfer feature information more effectively. To increase the flexibility of the model, we then use deformable convolution to deal with different nuclei irregular shapes and sizes. To reduce the predictive bias, we further construct multiple networks with different settings, which form an ensemble model. The proposed segmentation framework has achieved state-of-the-art accuracy on Herlev dataset with Zijdenbos similarity index (ZSI) of 0.933, and has the potential to be extended for solving other medical image segmentation tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge